
Long non-coding RNA (LncRNA) CHROMR promotes the expression of the CNNM1 gene by adsorbing hsa-miR-1299 to obtain drug resistance in diffuse large B lymphoma cells
Introduction
Diffuse large B-cell lymphoma (DLBCL) is a type of tumor composed of medium to large B lymphoid cells. The nucleus of tumor cells is equal to or exceeds that of normal macrophages; it is more than twice the size of the nucleus of normal lymphocytes and shows a diffuse growth pattern. DLBCL is the most common type of non-Hodgkin’s lymphoma (NHL) in hematological diseases, accounting for 30–40% of all cases of NHL. Of these, adult DLBCL cases in Western countries account for 25–35% of all NHL cases, but this can be as high as 45% in developing countries (1). DLBCL shows apparent heterogeneity in cell origin, morphology, immunohistochemical phenotype, molecular genetics, involved location, chemotherapy response, and survival rate (2). DLBCL can be a primary disease, or it can be transformed from other low-invasive lymphomas. The lesions mainly occur in the lymph nodes, and some can be found in the gastrointestinal tract, bones, and the central nervous system (3,4). In patients with DLBCL, the early lymph nodes enlarge rapidly, while the DLBCL outside the lymph nodes becomes tumor-like and can be accompanied by fibrosis. As DLBCL progresses, the lesions grow aggressively, leading to poor treatment effects (5,6). Although advances in medicine have promoted tremendous advances in the treatment of DLBCL, up to 40% of patients still die from relapse. At present, the molecular mechanism of DLBCL pathogenesis and drug resistance is still unclear, which seriously hinders the treatment of DLBCL (7,8).
Long non-coding RNA (lncRNA) is involved in many aspects of cell metabolism, such as cell tumorigenesis, proliferation, apoptosis, and drug resistance (9-13). Many lncRNAs, such as MALAT1, H19, BANCR, and HOTAIR have been intensely studied and used in clinical diagnoses, which verified that lncRNAs are essential participants in cancer progression and can be used as new biomarkers in the early diagnosis and treatment of some cancers (14-17). However, the role of lncRNA in cancer remains to be explored. At present, some lncRNAs are considered essential factors for cancer to acquire drug resistance. BCAR4 is highly expressed in prostate cancer. It can bind to the promoter region of GLI2 and activate downstream genes of GLI2, making prostate cancer cells less sensitive to androgen stimulation (18). In contrast, lnc-SNHG17 releases CD51 by adsorbing miR-144, thereby enhancing the proliferation and migration of prostate cancer cells (19). lncRNA CCAT1 acts on miR-24-3p to up-regulate FSCN1 and makes cancer cells resistant to paclitaxel (20).
lncRNA also plays a vital role in the occurrence, progression, resistance, and prognosis of DLBCL (21-23). LINC00857 can promote the occurrence and progression of DLBCL (24). SNHG14 can make DLBCL immune escape and promote proliferation (25). We found that CHROMR was differentially expressed in different rituximab-resistant cell lines. Furthermore, the main research direction of lncRNA CHROMR currently focuses on cholesterol diseases. However, there are no reports on the role of lncRNA CHROMR in cancer. Therefore, this article examines the mechanism by which lncRNA CHROMR acquires drug resistance in DLBCL. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1087/rc).
Methods
Cell culture and treatment
Human DLBCL cell lines, including SU_DHL_4, SU_DHL_8 (ATCC, Virginia, USA), were cultured in RIMI-1640 medium (Thermo Fisher Scientific, Massachusetts, USA), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin with an atmosphere of 5% CO2 at 37 ℃. When necessary, 17 nM rituximab (MCE) was added to the medium to induce cell death.
Cell transfection
According to the manufacturer’s instructions, miR-1299 mimic and its negative control (NC) (100 nM, Guangzhou all-perfect Biological Technology Co., Ltd., Guangzhou, China), pcDNA3.1-CHROMR, and pcDNA-NC (50 nM, Guangzhou all -perfect Biological Technology Co., Ltd.) vectors were transfected into SU_DHL_4 cells using Lipofectamine™ 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Related functional tests were performed 48 hours after transfection. Psicheck2.0-CHROMR-WT and psicheck2.0-CHROMR-MUT were co-transfected with miR-1299 mimic and mimic NC into 293T cells, and the fluorescence intensity was detected 48 hours after transfection.
Psicheck2.0-CNNM1-WT and psicheck2.0-CNNM1-MUT were co-transfected with miR-1299 mimic and mimic NC into 293T cells, and the fluorescence intensity was detected 48 hours after transfection.
Flow cytometry analysis for apoptosis
Cell apoptosis was assessed by flow cytometry using the Annexin V-FITC Apoptosis Detection Kit (Beyotime, Shanghai, China). Cells were collected, washed with phosphate buffered saline (PBS), and then stained with FITC-conjugated anti-Annexin V antibody and propidium iodide (PI) according to the manufacturer’s protocol. The percentage of Annexin V+ PI+ cells was determined with a flow cytometer (Becton Dickinson, New Jersey, USA).
Flow cytometry analysis for cell cycle
Cell cycle was assessed with flow cytometry using the Cell Cycle and Apoptosis Analysis Kit (Beyotime, Shanghai, China). Cells containing only the medium were used for the control. The cells were collected and then fixed in 70% ice-cold ethanol at 4 ℃ overnight. Cells were rewashed with cold PBS and suspended in PBS containing 0.1mg/mL RNase A and 50 µg/mL PI for 30min at room temperature. DNA contents of cells were determined with flow cytometry within an hour. Cell cycles were resolved by NovoExpress software (Agilent Technologies, Inc., CA, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The RNAiso plus kit (Takara, Dalian, China) was used to extract total RNA from SU_DHL_4, SU_DHL_8. The total RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara). RT-qPCR was performed on a LightCyclerR 96 PCR machine (Roche Diagnostics, Basel, Switzerland) using the SYBR-Green PCR kit (Takara Bio, Kusatsu, Japan). The reaction conditions were pre-denaturation at 95 ℃ for 10 min, denaturation at 95 ℃ for 10 sec, annealing at 60 ℃ for 20 sec, and extension at 72 ℃ for 34 sec, a total of 40 cycles. U6 small nuclear RNA (U6) or glyceraldehyde-3-phosphatedehydrogenase (GAPDH) served as the internal reference. Quantitative expression was calculated using the 2−ΔΔCT method (16). The primers used are shown in Table 1.
Table 1
| Primer | Sequences (5'-3') | Accession |
|---|---|---|
| CHROMR | F: TTCCCTGCACTCCAAACACAGA | NR_110204.1 |
| R: AAGTCTTCCATCTGTCAGCGCC | ||
| Hsa-miR-1299 | F: TTCTGGAATTCTGTGTGAGGGA | MIMAT0005887 |
| R: TTTTTTTTTTTTTTTTTTTTTTTT | ||
| GAPDH | F: ATGGTTTACATGTTCCAATATGA | NM_001256799 |
| R: TTACTCCTTGGAGGCCATGTGG | ||
| U6 | F: CGCTTCGGCAGCACATATAC | NR_004394 |
| R: AATATGGAACGCTTCACGA | ||
| CNNM1 | F: CTGCGCCAGGAGATAAGCA | NM_020348.3 |
| R: CCAGGTCACTGTAGGGGTCT |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Western blot analysis
The cells were lysed in Radioimmunoprecipitation assay (RIPA) buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 5 mM EDTA, 0.1 mM PMSF, and 2 mg/mL aprotinin. Protein concentration was measured using the BCA kit (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). Then, the proteins (50 µg/lane) were separated with 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (Beyotime Biotech, Shanghai, China) and transferred onto nitrocellulose membranes. The membranes were blocked with 3% blocking buffer (Beyotime) for 1 hour at room temperature and cultured with the primary antibodies CNNM1 (1:1,000, ab122648, Abcam), BAX (1:1,000, ab32503, Abcam), bate-actin (1:1,000, ab6276, Abcam), and BCL2 (1:1,000, ab32124, Abcam) overnight at 4 ℃. Subsequently, the membranes were cultured with the secondary antibody horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000, ab205718, Abcam) for 2 hours. Next, the membranes were visualized using an enhanced chemiluminescence reagent (Millipore). Protein blotting was analyzed using the Image Pro Plus 6.0 software (National Institutes of Health), with bate-actin as the internal reference. The experiment was repeated three times.
Dual-luciferase reporter gene assay
After 48 hours of transfection, the cell culture medium was discarded, the cell culture was washed with PBS once, then 200 µL of reporter gene cell lysate was added, the cells were fully lysed, centrifuged at 10,000–15,000 g for 3–5 minutes, and the supernatant was saved. The detection method is to add 65 µL of supernatant to a 96-well plate, add 100 µL of firefly luciferase detection reagent to each well for luminescence detection, and add 100 µL of renin luciferase detection reagent again for detection. The fluorescence detection value of Renilla was used as the benchmark to calibrate the detection value of firefly. The differences in luminous values of each group were compared.
Statistical analysis
All data were processed using the IBM SPSS Statistics version 21.0 software (IBM Corp., NY, USA). Data were first verified to show normal distribution and homogeneity of variance. Data were expressed as means ± standard deviation or counts. An independent samples t-test was used to compare two groups. P<0.05 indicated a statistically significant difference.
Results
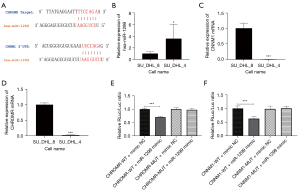
CHROMR is highly expressed in rituximab resistant DLBCL cell lines
Through bio-information analysis, we found that CHROMR was significantly highly expressed in rituximab-resistant cell lines. We predicted the targets of CHROMR and miR-1299 and CNNM1 and miR-1299 in Starbase and speculated a CHROMR/miR-1299/CNNM1 pathway related to the acquisition of drug resistance in DLBCL cell lines (Figure 1A). We selected rituximab sensitive DLBCL cell lines (SU_DHL_4) and rituximab resistant DLBCL cell lines (SU_DHL_8) for RT-qPCR detection. CHROMR and CNNM1 were found to be significantly high in SU_DHL_8, and miR-1299 was low in SU_DHL_8 (all P<0.001; Figure 1B-1D). Furthermore, we constructed a vector for the dual-luciferase target verification experiment. The results showed that miR-1299 reduced the firefly expression of psicheck2.0-CHROMR-WT and psicheck2.0-CNNM1-WT, while psicheck2.0-CHROMR-MUT and psicheck2.0-CNNM1-MUT were not affected (Figure 1E,1F). This shows that CHROMR and CNNM1 had targets for miR-1299. We found that CHROMR, miR-1299, and CNNM1 had significant differences in expression in cells with different sensitivity to rituximab, and there was a correlation between these three.
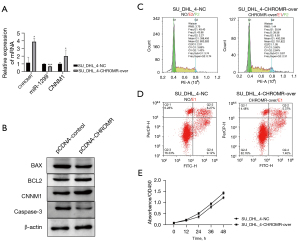
Overexpression of CHROMR increases the resistance of DLBCL cells to rituximab
To test our hypothesis, the pcDNA3.1-CHROMR vector was transfected into SU_DHL_4 cells and treated with 17 nM rituximab. We used RT-qPCR and WB to detect the expression of CHROMR, miR-1299, and CNNM1 before and after transfection. The expression of CHROMR and CNNM1 increased significantly (P<0.05), and the expression of miR-1299 decreased significantly (P<0.001; Figure 2A,2B). Subsequently, we tested with flow cytometry and found that, under the condition of 17 nM rituximab treatment, after the cells overexpressed CHROMR, the proportion of cells in the G2 phase increased by about three times that before overexpression, while apoptosis decreased by 6% (P<0.001; Figure 2C,2D). We also detected the expression of apoptosis-related proteins BAX, BCL2, and caspase-3 with WB and found that the expression of BAX and caspase-3 was significantly reduced (P<0.05) and the expression of BCL2 was significantly increased (P<0.01; Figure 2B). In addition, we also transfected SU_DHL_4 cells with pcDNA3.1-CHROMR vector and cultured them as usual. Through the cck8 experiment, we found that, after cells overexpress CHROMR, the level of cell proliferation increased (P<0.01; Figure 2E). These results show that CHROMR can increase the resistance of DLBCL cells to rituximab by regulating the cell cycle to promote cell proliferation and reduce the level of apoptosis.
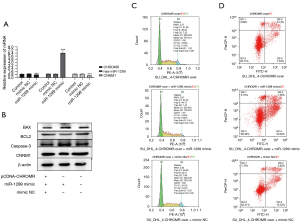
CHROMR/miR-1299/NCAM1 signaling pathway reduces the sensitivity of DLBCL cells to rituximab
We determined that CHROMR can enhance the resistance of DLBCL cells to rituximab, and we have also confirmed that there are interaction targets among CHROMR, miR-1299, and CNNM1. Therefore, does CHROMR regulate the cell cycle and apoptosis level of DLBCL cells through the CHROMR/miR-1299/CNNM1 pathway? We transfected pCDNA-CHROMR, pCDNA-CHROMR and miR-1299 mimic mixture, pCDNA-CHROMR, and mimic NC mixture into SU_DHL_4 cells, respectively, gave them 17 nM rituximab stimulation, and tested them 48 hours later. We confirmed that pCDNA-CHROMR and miR-1299 played a role in the cell through qPCR and WB and confirmed that there was a CHROMR/miR-1299/CNNM1 pathway (Figure 3A,3B). We then used flow cytometry to detect changes in the cell cycle and the level of apoptosis. When using the simple CHROMR overexpression cell group as the control group, we found that miR-1299 mimic increased the apoptosis level of CHROMR overexpression SU_DHL_4 cells (P<0.01) and reduced the proportion of cells in the G2 phase (P<0.01), which prevented cells from undergoing the proliferation cycle, but mimic NC had no such effect (Figure 3C,3D). The above results show that increasing the expression of miR-1299 can eliminate the rituximab resistance caused by the high expression of CHROMR. Results also suggest that the CHROMR/miR-1299/CNNM1 pathway can regulate the rituximab resistance of cells.
Discussion
In cancer, cholesterol levels were confirmed to be related to cancer progression. Studies showed that cholesterol levels in cancer cells are often higher than normal cells. There have been many studies in this area. In terms of epidemiological studies, studies showed that, for every 10 mg/dL increase in cholesterol levels, the recurrence rate of prostate cancer increased by 9%, and the use of lipid-lowering drugs (statins) is related to colorectal cancer mortality. In addition, there is a link between a slight reduction in cancer-related mortality of different cancer types. Still, some studies indicate that there is no link between cholesterol and cancer (26-29). In terms of the molecular mechanism, a study showed that, as cholesterol levels increased, the ability of anti-tumor cells TC9 cells polarized by CD8+ T cells to secrete IL-9 was impaired because cholesterol binds to LXRs and reduces the binding of LXRs Sumoylation to P65 and IL-9 promoter sequences (30). Therefore, in addition to acting on immune cells and weakening the lethality of tumor cells, does cholesterol affect tumor cells themselves? In this paper, through GEO analysis, we obtained the lncRNA difference data between rituximab-resistant DLBCL cells and rituximab-sensitive DLBCL cells. We found that the cholesterol-related lncRNA CHROMR was significantly high in expression. CHROMR is cholesterol-induced and related to metabolic regulation. The current research on lncRNA is mainly focuses on atherosclerosis-related diseases. CHROMR has been confirmed to be elevated in the plasma and atherosclerotic plaques of patients with coronary artery disease and is positively correlated with cholesterol levels (31). What role does CHROMR play in tumors, especially in the production of rituximab resistance in tumor cells? We overexpressed lncRNA CHROMR in rituximab-sensitive DLBCL cells SU_DHL_4 and then stimulated it with rituximab. We found that, under the killing effect of rituximab, compared with SU_DHL_4 cells that generally express CHROMR, SU_DHL_4, cells with high CHROMR expression have more cells to enter the cell proliferation cycle, the proliferation ability is enhanced, and the level of apoptosis is reduced. The results indicate that lncRNA CHROMR may reduce the killing effect of rituximab on tumor tissues by promoting cell proliferation.
We found the differential miRNA data of DLBCL through the HMDD v3.2 database, used the RegRNA database to predict the potential relationship between them and CHROMR, and found that miR-1299 and CHROMR have potential targets. Hsa-miR-1299 is a tumor suppressor, which has been confirmed in many cancers (32). miR-1299 can act on the EGFR/PI3K/AKT signaling pathway to inhibit non-small cell lung cancer development (33). miR-1299 can also act on NOTCH3 to make the ovarian cancer cell cycle stay in the G0G1 phase (34). In hepatocellular carcinoma, miR-1299 targets CDK6 to inhibit cell proliferation (35). But miR-1299 is not only a tumor suppressor; it is also associated with tumor drug resistance, reducing miR-1299 to promote paclitaxel resistance in ovarian cancer cells and breast cancer cells (36,37). In this study, we tested the expression of miR-1299 in different rituximab-resistant cell lines. miR-1299 was highly expressed in the rituximab-sensitive cell line SU_DHL_4. After CHROMR was overexpressed, the expression of miR-1299 decreased. Through the dual-luciferase experiment, we also verified that there was an interaction target between miR-1299 and CHROMR, and we confirmed that miR-1299 was the downstream target of CHROMR through the recovery experiment. We found that CHROMR makes DLBCL cells resistant to rituximab by adsorbing miR-1299.
CNNM1 is a cell cycle-related protein related to tumor development in prostate cancer and hepatocellular carcinoma and can regulate cell cycle and proliferation (38). In this study, we showed through experiments that CHROMR is overexpressed and CNNM1 is also overexpressed, but if the expression of miR-1299 is restored, the expression of CNNM1 will also be restored. Therefore, the result of this experiment is likely to constitute a ceRNA mechanism pathway with CHROMR and miR-1299. CHROMR increases the expression of CNNM1 by adsorbing miR-1299, thereby promoting the cycle progress of DLBCL cells and making DLBCL cells resistant to rituximab.
Currently, clinical samples suitable for detecting non-coding RNAs are patient blood, including serum, plasma, and blood leukocytes. Extracellular RNA is present in blood and can be rapidly quantified in a clinical setting. This property enables the clinical use of non-coding RNAs as biomarkers for diagnosing prognosis in diffuse large B lymphoma. However, the stability of lncRNA is poorer than that of miRNA, and its expression level is low, so it is challenging to develop it as a detection indicator, and it is mainly concentrated in experimental research at present. In addition, lncRNAs have tissue heterogeneity, and their expression varies significantly in different organs. Therefore, how to effectively target lncRNAs in specific organs is a problem that cannot be effectively solved. However, to explore the mechanism of action of lncRNA, we can intervene in lncRNA by interfering with the downstream of lncRNA, such as the CHROMR/miR-1299/CNNM1 pathway found in this experiment. We can try to judge by detecting the expression of miR-1299, The prognosis of patients, or increase the therapeutic effect of patients by inhibiting the expression of CNNM1.
Conclusions
Our study showed that there is a lncRNA CHROMR/miR-1299/CNNM1 pathway that makes DLBCL cells resistant to rituximab. This provides a new idea to solve rituximab resistance and better treat DLBCL.
Acknowledgments
Thanks to Guangzhou all-perfect Biological Technology Co., Ltd. for providing us with experimental technical guidance.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1087/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1087/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1087/coif). All authors report that Guangzhou all-perfect Biological Technology Co., Ltd provided the technological support of transfecting small fragments of RNA in cells. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Onaindia A, Santiago-Quispe N, Iglesias-Martinez E, et al. Molecular Update and Evolving Classification of Large B-Cell Lymphoma. Cancers (Basel) 2021;13:3352. [Crossref] [PubMed]
- Beham-Schmid C. Aggressive lymphoma 2016: revision of the WHO classification. Memo 2017;10:248-54. [Crossref] [PubMed]
- Small S, Barnea Slonim L, Williams C, et al. B Cell Lymphomas of the GI Tract. Curr Gastroenterol Rep 2021;23:9. [Crossref] [PubMed]
- Petrackova A, Turcsanyi P, Papajik T, et al. Revisiting Richter transformation in the era of novel CLL agents. Blood Rev 2021;49:100824. [Crossref] [PubMed]
- Hutchinson CB, Wang E. Primary mediastinal (thymic) large B-cell lymphoma: a short review with brief discussion of mediastinal gray zone lymphoma. Arch Pathol Lab Med 2011;135:394-8. [Crossref] [PubMed]
- Sugimoto KJ, Shimada A, Ichikawa K, et al. Primary platelet-derived growth factor-producing, spindle-shaped diffuse large B-cell lymphoma of the skull: a case report and literature review. Int J Clin Exp Pathol 2014;7:4381-90. [PubMed]
- Zhang XY, Collins GP, Cutter DJ, et al. Limited-stage diffuse large B-cell lymphoma: current management and challenges. Br J Haematol 2021;194:508-17. [Crossref] [PubMed]
- He MY, Kridel R. Treatment resistance in diffuse large B-cell lymphoma. Leukemia 2021;35:2151-65. [Crossref] [PubMed]
- Varier KM, Dhandapani H, Liu W, et al. An immunotherapeutic approach to decipher the role of long non-coding RNAs in cancer progression, resistance and epigenetic regulation of immune cells. J Exp Clin Cancer Res 2021;40:242. [Crossref] [PubMed]
- Di Fiore R, Suleiman S, Felix A, et al. An Overview of the Role of Long Non-Coding RNAs in Human Choriocarcinoma. Int J Mol Sci 2021;22:6506. [Crossref] [PubMed]
- Borkiewicz L, Kalafut J, Dudziak K, et al. Decoding LncRNAs. Cancers (Basel) 2021;13:2643. [Crossref] [PubMed]
- Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov 2021;20:629-51. [Crossref] [PubMed]
- Kai-Xin L, Cheng C, Rui L, et al. Roles of lncRNA MAGI2-AS3 in human cancers. Biomed Pharmacother 2021;141:111812. [Crossref] [PubMed]
- Seborova K, Vaclavikova R, Rob L, et al. Non-Coding RNAs as Biomarkers of Tumor Progression and Metastatic Spread in Epithelial Ovarian Cancer. Cancers (Basel) 2021;13:1839. [Crossref] [PubMed]
- Palanca-Ballester C, Rodriguez-Casanova A, Torres S, et al. Cancer Epigenetic Biomarkers in Liquid Biopsy for High Incidence Malignancies. Cancers (Basel) 2021;13:3016. [Crossref] [PubMed]
- Kidd SG, Carm KT, Bogaard M, et al. High expression of SCHLAP1 in primary prostate cancer is an independent predictor of biochemical recurrence, despite substantial heterogeneity. Neoplasia 2021;23:634-41. [Crossref] [PubMed]
- Zhao Z, Guo Y, Liu Y, et al. Individualized lncRNA differential expression profile reveals heterogeneity of breast cancer. Oncogene 2021;40:4604-14. [Crossref] [PubMed]
- Cai Z, Wu Y, Li Y, et al. BCAR4 activates GLI2 signaling in prostate cancer to contribute to castration resistance. Aging (Albany NY) 2018;10:3702-12. [Crossref] [PubMed]
- Bai M, Lei Y, Wang M, et al. Long Non-coding RNA SNHG17 Promotes Cell Proliferation and Invasion in Castration-Resistant Prostate Cancer by Targeting the miR-144/CD51 Axis. Front Genet 2020;11:274. [Crossref] [PubMed]
- Li X, Han X, Wei P, et al. Knockdown of lncRNA CCAT1 enhances sensitivity of paclitaxel in prostate cancer via regulating miR-24-3p and FSCN1. Cancer Biol Ther 2020;21:452-62. [Crossref] [PubMed]
- Zhou M, Zhao H, Xu W, et al. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer 2017;16:16. [Crossref] [PubMed]
- Karstensen KT, Schein A, Petri A, et al. Long Non-Coding RNAs in Diffuse Large B-Cell Lymphoma. Noncoding RNA 2020;7:1. [Crossref] [PubMed]
- Huang X, Qian W, Ye X. Long Noncoding RNAs in Diffuse Large B-Cell Lymphoma: Current Advances and Perspectives. Onco Targets Ther 2020;13:4295-303. [Crossref] [PubMed]
- Huang Y, Lin Y, Song X, et al. LINC00857 contributes to proliferation and lymphomagenesis by regulating miR-370-3p/CBX3 axis in diffuse large B-cell lymphoma. Carcinogenesis 2021;42:733-41. [Crossref] [PubMed]
- Tian Y, Li L, Lin G, et al. lncRNA SNHG14 promotes oncogenesis and immune evasion in diffuse large-B-cell lymphoma by sequestering miR-152-3p. Leuk Lymphoma 2021;62:1574-84. [Crossref] [PubMed]
- Bjerre LM, LeLorier J. Do statins cause cancer? A meta-analysis of large randomized clinical trials. Am J Med 2001;110:716-23. [Crossref] [PubMed]
- Allott EH, Howard LE, Cooperberg MR, et al. Serum lipid profile and risk of prostate cancer recurrence: Results from the SEARCH database. Cancer Epidemiol Biomarkers Prev 2014;23:2349-56. [Crossref] [PubMed]
- Nielsen SF, Nordestgaard BG, Bojesen SE. Statin use and reduced cancer-related mortality. N Engl J Med 2012;367:1792-802. [Crossref] [PubMed]
- Ravnskov U, Rosch PJ, McCully KS. Statins do not protect against cancer: quite the opposite. J Clin Oncol 2015;33:810-1. [Crossref] [PubMed]
- Ma X, Bi E, Huang C, et al. Cholesterol negatively regulates IL-9-producing CD8+ T cell differentiation and antitumor activity. J Exp Med 2018;215:1555-69. [Crossref] [PubMed]
- Hennessy EJ, van Solingen C, Scacalossi KR, et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat Metab 2019;1:98-110. [Crossref] [PubMed]
- Kaiyuan D, Lijuan H, Xueyuan S, et al. The role and underlying mechanism of miR-1299 in cancer. Future Sci OA 2021;7:FSO693. [Crossref] [PubMed]
- Cao S, Li L, Li J, et al. MiR-1299 Impedes the Progression of Non-Small-Cell Lung Cancer Through EGFR/PI3K/AKT Signaling Pathway. Onco Targets Ther 2020;13:7493-502. [Crossref] [PubMed]
- Pei Y, Li K, Lou X, et al. miR-1299/NOTCH3/TUG1 feedback loop contributes to the malignant proliferation of ovarian cancer. Oncol Rep 2020;44:438-48. [Crossref] [PubMed]
- Zhu H, Wang G, Zhou X, et al. miR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed Pharmacother 2016;83:792-7. [Crossref] [PubMed]
- Liu G, Zhang Z, Song Q, et al. Circ_0006528 Contributes to Paclitaxel Resistance of Breast Cancer Cells by Regulating miR-1299/CDK8 Axis. Onco Targets Ther 2020;13:9497-511. [Crossref] [PubMed]
- Xia B, Zhao Z, Wu Y, et al. Circular RNA circTNPO3 Regulates Paclitaxel Resistance of Ovarian Cancer Cells by miR-1299/NEK2 Signaling Pathway. Mol Ther Nucleic Acids 2020;21:780-91. [Crossref] [PubMed]
- Xie Y, Wang Y, Gong R, et al. SNHG7 Facilitates Hepatocellular Carcinoma Occurrence by Sequestering miR-9-5p to Upregulate CNNM1 Expression. Cancer Biother Radiopharm 2020;35:731-40. [Crossref] [PubMed]
(English Language Editor: C. Mullens)


