
Analysis and prediction model of ferroptosis related genes in breast cancer
Introduction
Breast cancer (BRCA) is the main cause of cancer-related death in women. The latest data show that approximately 2.1 million new occurrences of BRCA were diagnosed in 2018, accounting for 25% of all female cancers (1). In China, BRCA ranks first in the incidence of malignant tumors in females; its mortality rate ranks fifth and has been increasing in recent years (2). As treatment methods continue to improve, molecular classification has changed the pattern of treatment. According to tumor stage, grade, human epidermal growth factor receptor 2, hormone receptor status, and tumor proliferation index, molecular classification provides a reference for clinical treatment selection and prognosis evaluation (3,4). BRCA is a highly heterogeneous tumor; different molecular types have different prognosis, and there are also differences in the same molecular type (5). Therefore, it is important to explore new treatment directions to improve survival prediction performance.
Selective induction of cancer cell death is one of the most effective treatment methods for tumors. One such method is ferroptosis, which is closely related to various diseases, especially cancer, and plays a key role in all aspects of cancer biology and drug resistance (6). Ferroptosis is a new type of non-apoptosis regulated cell death that inhibits or promotes tumor progression by releasing a variety of signaling molecules in the tumor microenvironment (7). A Study have confirmed that p53 promotes tumor cell ferroptosis through regulation and reduces the probability of distant tumor metastasis (8). Ferroptosis also affects tumor resistance and cancer immunotherapy (9,10). Recently, one study has shown that triple-negative BRCA is more sensitive to ferroptosis than hormone receptor-positive BRCA, which suggests that ferroptosis could be an effective treatment strategy for triple-negative BRCA (11). However, there remains a need for more extensive research on the application of ferroptosis towards the treatment of BRCA. Therefore, this study aimed to explore the mechanism and biological behavior of ferroptosis by obtaining information on BRCA patients from The Cancer Genome Atlas (TCGA) database. We also sought to identify potential treatment strategies for BRCA. We present the following article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2686/rc).
Methods
Data collection
RNA-sequencing expression (level 3) profiles and corresponding clinical information for BRCA were downloaded from TCGA dataset (https://portal.gdc.cancer.gov/legacy-archive/search/f). A total of 113 paracancerous samples were collected from TCGA database and 54 samples were collected from non-diseased tissue sites in GTEx.
Bioinformatics analysis
We used R (v4.0.3) software packages “ggplot2” and “pheatmap” to plot 25 ferroptosis-related genes (CDKN1A, HSPA5, EMC2, SLC7A11, NFE2L2, MT1G, HSPB1, GPX4, FANCD2, CISD1, FDFT1, SLC1A5, SAT1, TFRC, RPL8, NCOA4, LPCAT3, GLS2, DPP4, CS, CARS1, ATP5MC3, ALOX15, ACSL4, and ATL1). This was done to create heat maps of differential expression in BRCA and normal control samples and to conduct correlation analysis. Ferroptosis-related genes were derived from a systematic analysis of the abnormalities and functions of ferroptosis in cancer by Liu et al. (12).
Survival analysis
Kaplan-Meier (KM) curves were drawn using R software packages “survival” and “survminer”. Log-rank was used to test the KM survival analysis and to compare the survival difference of the above-mentioned ferroptosis-related genes in BRCA; the P value (P<0.05) was considered statistically significant.
Prognostic model establishment
A LASSO regression algorithm was used to select the characteristics of BRCA ferroptosis-related genes using 10-fold cross-validation; log-rank was used to test the KM survival analysis and compare the survival differences between the two groups of high and low wind directions. Lastly, time receiver operating characteristic (ROC) analysis was conducted to compare the accuracy of the predicted sex and risk scores for five genes.
Establishment of a nomogram
Uni-factor and multi-factor Cox regression analysis was performed using forest plots through the “forestplot” R package to display the P value, hazard ratio, and 95% confidence interval of each variable. Based on the results of the multivariate Cox proportional hazard analysis, the R software package “rms” was used to establish a nomogram to predict 3–5 years of disease-specific survival.
Statistical analysis
Bioinformatics analyses of databases were described in detail in the above method. The two-sided Student’s t-test was used to compare the differences between groups. KM analysis was performed via KM plotter. All assays were performed using R (4.0.3). P<0.05 was considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). TCGA data disclosure was available for this study without the approval of the ethics committee.
Results
Expression of ferroptosis-related genes in BRCA
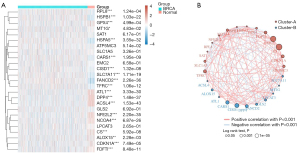
Through the analysis of 25 ferroptosis-related genes (Table S1), 19 ferroptosis-related genes (CDKN1A, HSPA5, SLC7A11, NFE2L2, MT1G, HSPB1, GPX4, FANCD2, CISD1, FDFT1, TFRC, RPL8, NCOA4, DPP4, CS, CARS1, ALOX15, ACSL4, and ATL1) showed differences in tumors with BRCA compared to those in normal tissues (Figure 1A). The genes related to ferroptosis in BRCA are closely related. The most significant positive correlations were between EMC2 and CS, and the most significant negative correlations were between CISD1 and CARS1 (Figure 1B).
Survival analysis and prognostic model establishment
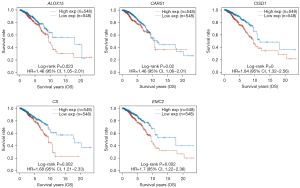
Survival analysis of the differentially expressed genes revealed that the five ferroptosis-related genes, CISD1, ALOX15, CS, CARS1, and EMC2, showed significant differences in survival in BRCA (P<0.05); there was no significant survival difference in the other ferroptosis-related genes (P<0.05; Figure 2). According to the expression profiles of the 19 ferroptosis-related genes obtained from the previous analysis, a prognosis model was constructed using LASSO Cox regression analysis. After regression analysis, the best four genes and their corresponding correlation coefficients were obtained.
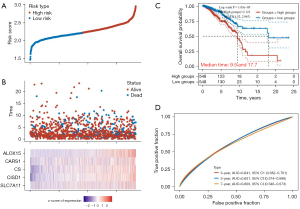
The risk score was calculated as follows: risk score = (0.361) × CISD1 + (0.176) × ALOX15 + (0.2704) × CARS1 + (0.1208) × SLC7A11. According to the median risk score, patients were divided into a high-risk group (548 cases) and a low-risk group (548 cases; Figure 3A). The BRCA survival rate of the high-risk group was significantly higher than that of the low-risk group (Figure 3B). The KM survival analysis curve showed that the survival rate of the low-risk group was significantly higher than that of the high-risk group (P<0.001; Figure 3C). The ROC curve was used to evaluate the predictive performance of the prognostic model. The area under the ROC curve was 0.641 at 3 years, 0.631 at 5 years, and 0.609 at 7 years (Figure 3D).
BRCA patient nomogram
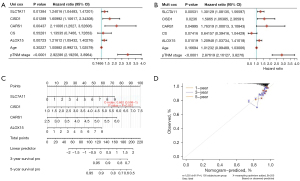
Univariate and multivariate Cox regression analyses of available clinical indicators, age, pTNM staging, and four ferroptosis-related genes were performed to determine independent prognostic predictors of disease-specific survival. Cox regression analysis showed that after adjusting for confounding factors, pTNM staging and CISD1, ALOX15, CARS1, and SLC7A11 scores could all be used as independent prognostic factors (Figure 4A,4B). Based on the ferroptosis-related gene risk factors, 3- and 5-year prognostic prediction nomograms of BRCA patients were drawn; the c-index was 0.662 and confidence interval was 0.598–1 (Figure 4C). Evaluation for consistency of the calibration curve showed that the survival rate of the 3- and 5-year prognosis prediction nomogram of patients is in good agreement with the actual survival rate (Figure 4D).
Discussion
As an inevitable biological event in organisms, cell death plays a vital role in the regulation of homeostasis and development. Ferroptosis, a regulated form of cell death, has different biochemical processes and genetic characteristics mainly driven by iron accumulation, lipid peroxidation, and plasma membrane rupture (13). Compared to normal cells, cancer cells exhibit iron ion aggregation, and regulating ferroptosis from the perspective of iron homeostasis can effectively kill tumor cells (14). In recent years, studies have confirmed that using ferroptosis to induce cancer cell death is effective for advanced tumors and tumor resistance (15,16). In addition to various inducing molecules, many genes can be used as markers of ferroptosis (17). Ferroptosis-related genes have shown good predictive performance in many tumors. BRCA has the highest incidence of malignant tumors in women; however, there are few studies on using ferroptosis-related genes to predict the prognosis of BRCA.
To address this need, we obtained clinical BRCA gene expression data from TCGA database and selected ferroptosis-related genes through a literature search. Here, we analyzed the correlation between ferroptosis-related genes and BRCA through differential gene analysis and determined that 19 ferroptosis-related genes were closely related to BRCA. Survival analysis of these differential genes revealed that four ferroptosis-related genes, CISD1, ALOX15, CARS1, and SLC7A11, had significant differences in survival in BRCA. The survival of the high-expression group was significantly lower than that of the low-expression group. LASSO Cox regression analysis was used to construct a prognostic model which divided BRCA patients into high- and low-risk groups. We observed that patients in the low-risk group had longer survival times than those in the high-risk group. In addition, we developed a nomogram, ROC curves, and calibration charts based on the results of the multivariate Cox regression to confirm the predictive ability of the nomogram.
Finally, a risk-scoring model consisting of four ferroptosis-related genes, CISD1, ALOX15, CARS1, and SLC7A11, was constructed. As a mitochondrial protein coding gene, CISD1 is an important regulator of ferroptosis. Its anti-ferroptosis activity is closely related to changes in the mitochondrial iron accumulation. CISD1 expression is associated with tumor growth and is a potential therapeutic target (18). ALOX15 may also be a potential target for cancer treatment, and its expression level can be used as a predictive marker for the clinical outcome of patients with lymph node metastasis and malignant tumors (19). CARS1 participates in cysteine metabolism and is the rate-limiting agent in glutathione synthesis. Glutathione is an important molecule that can be used as a predictor of ferroptosis in kidney cancer, and it regulates the oxidative environment of cells and ferroptosis (20). SLC7A11 expression is associated with amino acid transport essential in glutathione homeostasis and protection of cells from oxidative stress; it also plays a key role in oxidative stress signals that are closely related to cell proliferation and tumor growth (21).
In summary, we used bioinformatic analyses to construct a new BRCA prognostic model that contains four ferroptosis-related genes which can independently predict the overall survival of patients with BRCA. Additionally, a nomogram was developed to accurately predict the prognosis of BRCA. However, there are limitations to the study because the research data came from a database lacking some clinical information. Furthermore, the specific mechanism of the relationship between ferroptosis-related genes and BRCA prognosis needs further study.
Acknowledgments
Funding: This study was supported by Chongqing Science and Health Joint Medical Research Project (No. 2019QNXM027).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2686/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2686/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2686/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2686/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Li H, Zheng RS, Zhang SW, et al. Incidence and mortality of female breast cancer in China, 2014. Zhonghua Zhong Liu Za Zhi 2018;40:166-71. [PubMed]
- Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA 2019;321:288-300. [Crossref] [PubMed]
- Liang Y, Zhang H, Song X, et al. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin Cancer Biol 2020;60:14-27. [Crossref] [PubMed]
- Denkert C, von Minckwitz G, Darb-Esfahani S, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 2018;19:40-50. [Crossref] [PubMed]
- Valashedi MR, Najafi-Ghalehlou N, Nikoo A, et al. Cashing in on ferroptosis against tumor cells: Usher in the next chapter. Life Sci 2021;285:119958. [Crossref] [PubMed]
- Jiang M, Qiao M, Zhao C, et al. Targeting ferroptosis for cancer therapy: exploring novel strategies from its mechanisms and role in cancers. Transl Lung Cancer Res 2020;9:1569-84. [Crossref] [PubMed]
- Viswanathan VS, Ryan MJ, Dhruv HD, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 2017;547:453-7. [Crossref] [PubMed]
- Wang W, Green M, Choi JE, et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569:270-4. [Crossref] [PubMed]
- Zheng DW, Lei Q, Zhu JY, et al. Switching Apoptosis to Ferroptosis: Metal-Organic Network for High-Efficiency Anticancer Therapy. Nano Lett 2017;17:284-91. [Crossref] [PubMed]
- Li Z, Chen L, Chen C, et al. Targeting ferroptosis in breast cancer. Biomark Res 2020;8:58. [Crossref] [PubMed]
- Liu Z, Zhao Q, Zuo ZX, et al. Systematic Analysis of the Aberrances and Functional Implications of Ferroptosis in Cancer. iScience 2020;23:101302. [Crossref] [PubMed]
- Liu J, Kuang F, Kroemer G, et al. Autophagy-Dependent Ferroptosis: Machinery and Regulation. Cell Chem Biol 2020;27:420-35. [Crossref] [PubMed]
- Lei P, Ayton S, Bush AI. The essential elements of Alzheimer's disease. J Biol Chem 2021;296:100105. [Crossref] [PubMed]
- Xu G, Wang H, Li X, et al. Recent progress on targeting ferroptosis for cancer therapy. Biochem Pharmacol 2021;190:114584. [Crossref] [PubMed]
- Sui S, Xu S, Pang D. Emerging role of ferroptosis in breast cancer: New dawn for overcoming tumor progression. Pharmacol Ther 2022;232:107992. [Crossref] [PubMed]
- Chen X, Comish PB, Tang D, et al. Characteristics and Biomarkers of Ferroptosis. Front Cell Dev Biol 2021;9:637162. [Crossref] [PubMed]
- Yuan H, Li X, Zhang X, et al. CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem Biophys Res Commun 2016;478:838-44. [Crossref] [PubMed]
- Fochtmann-Frana A, Haymerle G, Schachner H, et al. Expression of 15-lipoxygenase-1 in Merkel cell carcinoma is linked to advanced disease. Clin Otolaryngol 2018;43:1335-44. [Crossref] [PubMed]
- Wu G, Wang Q, Xu Y, et al. A new survival model based on ferroptosis-related genes for prognostic prediction in clear cell renal cell carcinoma. Aging (Albany NY) 2020;12:14933-48. [Crossref] [PubMed]
- Ma Z, Zhang H, Lian M, et al. SLC7A11, a component of cysteine/glutamate transporter, is a novel biomarker for the diagnosis and prognosis in laryngeal squamous cell carcinoma. Oncol Rep 2017;38:3019-29. [Crossref] [PubMed]


