
Clinical significance of ALKBH4 expression in non-small cell lung cancer
Introduction
Globally, lung cancer is the second most commonly diagnosed cancer next to the breast cancer and is the highest cause of cancer deaths (1). The reasons for the high mortality of non-small cell lung cancer (NSCLC) are that patients are diagnosed at an advanced stage and recurrence is common, even among patients who receive curative surgery. Although the prognosis of NSCLC patients has improved with the advent of molecular targeted drugs, such as tyrosine kinase inhibitors against epidermal growth factor receptor (EGFR) mutations (2) and echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML-ALK) fusion gene mutations (3) and immune checkpoint inhibitors (4), it is difficult to cure NSCLC with drugs alone. Thus, novel molecular markers for the diagnosis and prediction of prognosis are required to select appropriate treatment for patients with NSCLC.
Gene methylation is deeply involved in epigenetics and affects not only development and maintenance of homeostasis but also carcinogenesis. Escherichia coli AlkB is a 2-oxoglutarate and Fe (II)-dependent dioxygenase that repairs alkylated DNA/RNA nucleotides by catalysing oxidative demethylation (5-7). In humans, the existence of nine AlkB homolog (ALKBH) family members (ALKBH1–9) has been reported (8,9). Some of the ALKBH molecules recognise various types of substrates, including methylated single- or double-stranded DNA or RNA bases (10). Recently, it has been reported that the expression of ALKBH affects the progression of several cancers. The high expression of ALKBH3 and ALKBH5 in NSCLC was reported to be associated with a poor prognosis (11,12). ALKBH4 is one of the ALKBH family proteins. Li et al. (13) demonstrated that dioxygenase ALKBH4-mediated demethylation of a monomethylated site in actin (K84me1) regulates actin-myosin interaction and actomyosin-dependent processes such as cytokinesis and cell migration. According to our previous experimental study with NSCLC cells, ALKBH4 was suggested to play a part in cell proliferation via the upregulation of the expression of the multifunctional oncogene E2 promoter-binding factor 1 (E2F1) (14). Unfortunately, the role of ALKBH4 in clinical characters and the prognostic outcome of NSCLC is poorly understood because the previous study analyzed selected patients with large adenocarcinoma. In addition, because the previous study only evaluated the results by a univariate analysis, the independent predictive value of ALKBH4 in determining the prognostic outcome remains unclear. In the current study, we comprehensively evaluated patients with NSCLC without any selection criteria regarding tumor size and histologic subtypes by univariate and multivariate analyses. We also reviewed previous reports and considered the role of the ALKBH family expression in various cancers. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-39/rc).
Methods
Patients and specimens
This retrospective cohort study enrolled 160 NSCLC patients who were completely resected at Kagoshima University Hospital from January 2001 to December 2007 (Table 1). The patients received lobectomy in 154 cases, bi-lobectomy in 4 cases, and pneumonectomy in 2 cases with mediastinal lymphadenectomy. In this study, cases receiving induction therapy before operation were not included. The average age of 160 NSCLC patients (102 males and 58 females) who were enrolled in this study was 69 years (range, 26–84 years). Based on the results of pathological examination using surgical specimens, the histological type was lung adenocarcinoma (LUAD) (n=112), lung squamous cell carcinoma (LUSC) (n=40) or other (n=8) including large cell carcinoma, adenosquamous carcinoma, mucoepidermoid carcinoma, pleomorphic carcinoma. According to the Association for the Study of Lung Cancer TNM (tumor-node-metastasis) classification, 7th edition (15), progression level of NSCLC was classified into stages IA (n=50), IB (n=52), IIA (n=16), IIB (n=15), and IIIA (n=27). This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Committee of Kagoshima University Hospital (registration number 351) and informed consent was taken from all the patients.
Table 1
| Characteristics | Value |
|---|---|
| Sex | |
| Male | 102 |
| Female | 58 |
| Age | |
| Mean, years [range] | 69±9 [26–84] |
| Operation | |
| Pneumonectomy | 2 |
| Bilobectomy | 4 |
| Lobectomy | 154 |
| Stage | |
| IA | 50 |
| IB | 52 |
| IIA | 16 |
| IIB | 15 |
| IIIA | 27 |
| Histology | |
| LUAD | 112 |
| LUSC | 40 |
| Others | 8 |
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Immunohistochemistry of ALKBH4 in NSCLC
After the surgery, the specimens were fixed with formalin and embedded in paraffin. Then, they were cut into 3 µm thick sections and mounted on a glass slide for immunohistochemistry. Next, the samples were deparaffinized with xylene and gradually dehydrated with an ethanol series. The slides were immersed in a 0.3% hydrogen peroxide solution in methanol for 30 minutes at room temperature to block the endogenous peroxidase activity. Sections were washed 3 times with phosphate buffered saline (PBS) for 5 minutes each and then treated with 1% bovine serum albumin for 30 minutes at room temperature to block non-specific reactions. The blocked sections were incubated with the rabbit polyclonal antibody against human ALKBH4 (1:200; Novus Biologicals, Littleton, CO, USA, NBP2-14737) and left overnight at 4 ℃, followed by staining with a streptavidin-biotin peroxidase kit (Vector Laboratories, Inc., Burlingame, CA, USA). The sections were washed in PBS for 5 minutes three times and the immune complex was visualized by incubating sections with diaminobenzidine tetrahydrochloride. The sections were rinsed briefly in water, counterstained with hematoxylin, and mounted. Noncancerous colon samples were used as positive controls for ALKBH4. The ALKBH4 expression was determined by counting the number of cancer cells in which the cytoplasm was stained with anti-ALKBH4 antibodies. Two investigators independently evaluated the ALKBH4 expression via immunohistochemistry within each tumor by assessing a total of 1,000 cancer cells in 10 selected fields (100 cells/field) using high-power (×200) microscopy. The average labeling index of ALKBH4 was assessed according to the proportion of positive cells in each field. The ALKBH4 expression was graded as ALKBH4-positive if >20% of cancer cells were stained or as ALKBH4-negative if <20% of cancer cells were stained.
Statistical analysis
Group differences were compared using the chi-squared test. Factors associated with overall survival (OS) and recurrence-free survival (RFS) were assessed by univariate and multivariate logistic regression analyses. Clinical factors included age, sex, tumor size, pl, pm, pT, pN, ALKBH4. Because of the relatively limited number of ALKBH4 positive cases, we used propensity score adjustment for each of the above risk factors for multivariable modeling. Propensity score adjustment preserved statistical power by reducing covariates into a single variable. For example, when the adjusted effect of ALKBH4 for survival was evaluated, the propensity score was created through a binary logistic regression providing the predicted probability of having ALKBH4 as a function of the other candidate risk factors (age, sex, tumor size, pl, pm and pN for RFS, and tumor size, pm and pN for OS). Propensity scores were computed separately for each candidate risk factor and then used as a covariate in the model adjusted for the effect of each factor. P values of <0.05 were considered statistically significant. Data were analyzed using the Statistical Package for the Social Sciences (SPSS version 26.0).
Results
The ALKBH4 expression and clinicopathological factors
In the NSCLC clinical samples, the expression of was identified not only in cell membranes but also in the cytoplasm of cancer cells (Figure 1). In 140 of 160 cases, ALKBH4 was more highly expressed in the cancerous tissue than in the surrounding normal tissue. The average expression rate of ALKBH4 in all cases was 16.3%. According to the immunohistochemical evaluation of the samples, the patients were classified into an ALKBH4-positive group (n=58, 36%) and an ALKBH4-negative group (n=102). Table 2 shows the expression of ALKBH4 and the correlation with clinicopathological factors. Regarding histological type, ALKBH4 was more highly expressed in LUAD than in other types (P=0.011). Moreover, we classified the intensity of the ALKBH4 expression in each cancer cell of LUAD and LUSC as strong, moderate, weak, or none (Figure 1D-1K). The expression intensity of ALKBH4 in each cancer cell was also stronger in LUAD than in LUSC (Table 3). Furthermore, we investigated the association between ALKBH4 and clinical factors in LUAD, but found no significant association with any of the factors. In this study, there were four cases of adenocarcinoma in situ, and the moderate expression of ALKBH4 was observed in two cases, while the weak expression ALKBH4 was observed in the remaining two cases (Figure 1C).
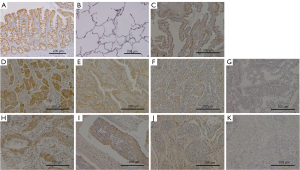
Table 2
| Clinical factors | Expression of ALKBH4 | P value | |
|---|---|---|---|
| Positive (n=58, 36%) | Negative (n=102, 64%) | ||
| Age, years | 0.513 | ||
| <70 | 24 (33%) | 48 (67%) | |
| ≥70 | 34 (39%) | 54 (61%) | |
| Sex | 0.123 | ||
| Male | 32 (31%) | 70 (69%) | |
| Female | 26 (45%) | 32 (55%) | |
| Tumor size, mm | >0.999 | ||
| <30 | 29 (37%) | 50 (63%) | |
| ≥30 | 29 (36%) | 52 (64%) | |
| pl | 0.736 | ||
| Yes | 22 (38%) | 36 (62%) | |
| No | 36 (35%) | 66 (65%) | |
| pm | 0.800 | ||
| Yes | 7 (39%) | 11 (61%) | |
| No | 51 (36%) | 91 (64%) | |
| pT | >0.999 | ||
| T1 | 20 (36%) | 35 (64%) | |
| ≥T2 | 38 (36%) | 67 (64%) | |
| pN | >0.999 | ||
| Yes | 13 (36%) | 23 (64%) | |
| No | 45 (36%) | 79 (64%) | |
| Stage | 0.864 | ||
| I | 38 (37%) | 64 (63%) | |
| ≥II | 20 (34%) | 38 (66%) | |
| Histology | 0.011 | ||
| LUAD | 48 (43%) | 64 (57%) | |
| Others | 10 (21%) | 38 (79%) | |
ALKBH, AlkB homolog; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma.
Table 3
| ALKBH4 intensity | LUAD | LUSC | P value |
|---|---|---|---|
| Strong | 23 (20.5%) | 2 (5%) | 0.036 |
| Middle | 32 (28.6%) | 8 (20%) | |
| Weak | 44 (39.3%) | 24 (60%) | |
| None | 13 (11.6%) | 6 (15%) |
ALKBH, AlkB homolog; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Univariate and multivariate logistic regression analyses of survival
During the median follow-up of 70 months (range, 1–218 months), 86 relapse and 64 deaths occurred. According to a survival analysis using clinical NSCLC samples, the RFS in the ALKBH4-positive group tended to be poorer than that in the ALKBH4-negaive group (P=0.091; Figure 2A), although OS was not correlated with the expression of ALKBH4 (P=0.141; Figure 2B). When the analysis was restricted to LUAD patients, both RFS and OS rates were significantly lower in the ALKBH4-positive group than in the negative-group (RFS: P=0.008, Figure 2C; OS: P=0.031, Figure 2D). Tables 4,5 show the results of univariate and multivariate logistic regression analyses of factors related to RFS (Table 4) and OS (Table 5) of LUAD. The univariate analysis showed that age (P=0.030), sex (P=0.019), tumor size (P=0.010), pl (P=0.042), pm (P=0.005), p-T factor (P=0.038), p-N factor (P<0.001) and the expression of ALKBH4 (P=0.008) were significantly related to RFS. The multivariate analysis indicated that the ALKBH4 expression [odds ratio (OR) =2.234; 95% confidence interval (CI): 1.303–3.832; P=0.003] and p-N factor (OR =3.067; 95% CI: 1.689–5.556; P<0.001) were independent prognostic factors of RFS. Moreover, the univariate analysis showed that tumor size (P=0.009), pm (P=0.008), p-N factor (P<0.001) and the expression of ALKBH4 (P=0.031) were significantly related to OS. A multivariate analysis indicated that the expression of ALKBH4 (OR =2.324; 95% CI: 1.192–4.530; P=0.013) and the p-N factor (OR =3.236; 95% CI: 1.664–6.289; P=0.001) were independent prognostic factors for OS.
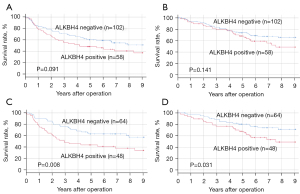
Table 4
| Clinical factors | Univariate P | Adjusted OR | 95% CI | P value |
|---|---|---|---|---|
| Age | 0.030 | 1.527 | 0.883–2.640 | 0.13 |
| Sex | 0.019 | 1.247 | 0.685–2.273 | 0.47 |
| Tumor size | 0.010 | 1.325 | 0.764–2.297 | 0.317 |
| pl | 0.042 | 1.388 | 0.750–2.570 | 0.297 |
| pm | 0.005 | 1.425 | 0.685–2.967 | 0.343 |
| pT | 0.038 | – | – | – |
| pN | <0.001 | 3.067 | 1.689–5.556 | <0.001 |
| ALKBH4 | 0.008 | 2.234 | 1.303–3.832 | 0.003 |
RFS, recurrence-free survival; LUAD, lung adenocarcinoma; OR, odds ratio; CI, confidence interval; ALKBH, AlkB homolog.
Table 5
| Clinical factors | Univariate P | Adjusted OR | 95% CI | P value |
|---|---|---|---|---|
| Age | 0.149 | – | – | – |
| Sex | 0.153 | – | – | – |
| Tumor size | 0.009 | 2.068 | 1.068–4.005 | 0.031 |
| pl | 0.713 | – | – | – |
| pm | 0.008 | 1.969 | 0.894–4.329 | 0.093 |
| pT | 0.585 | – | – | – |
| pN | <0.001 | 3.236 | 1.664–6.289 | 0.001 |
| ALKBH4 | 0.031 | 2.324 | 1.192–4.530 | 0.013 |
OS, overall survival; LUAD, lung adenocarcinoma; OR, odds ratio; CI, confidence interval; ALKBH, AlkB homolog.
Discussion
Our previous experimental study showed the preliminary results of the ALKBH4 expression in an immunohistochemical study of selected patients with large LUAD. Therefore, the relationship among tumor size and histologic subtype (LUAD vs. LUSC), the expression of ALKBH4 protein, and the prognostic outcome remained unclear (14). In the current study, we comprehensively evaluated the ALKBH4 expression in NSCLC with a multivariate analysis without any selection criteria. As a result, the ALKBH4 expression was not associated with tumor size, but was associated with histological type. According to a stratification analysis, the expression of ALKBH4 was significantly associated with a worse prognosis in patients with LUAD, but not in patients with LUSC. These results may suggest that ALKBH4 plays an important role in the development and progression of LUAD.
The expression status of ALKBH family mRNA or protein in various cancers is summarized in Table 6. The expression of the ALKBH family was upregulated in most cancers (11,12,16-20,23,25-27), but the expression of ALKBH1 protein in gastric cancer (16), ALKBH5 and 9 mRNA in clear cell renal cell cancer (RCC) (22), and ALKBH5 mRNA in colon cancer (24) were downregulated. Interestingly, it has been reported that the ALKBH1 mRNA and protein expression levels differ in gastric cancer (16). According to our previous study, the ALKBH4 mRNA and protein expression levels were higher in LUAD and LUSC than in normal tissues (14). The current study revealed that the expression of ALKBH4 protein was also higher in NSCLC than in normal tissues. With regard to the expression rate and intensity of ALKBH4, the proportion of ALKBH4-positive cells was higher in LUAD than in other types. Likewise, the expression intensity of ALKBH4 protein in each tumor cell was stronger in LUAD than in LUSC. According to these findings, together with the prognostic outcome, the expression of ALKBH4 protein is considered to have more important implications in LUAD in comparison to LUSC.
Table 6
| ALKBH subtype | Cancer | Target factor | Expression of cancer tissue | Correlation with clinical factors | Cause of poor prognosis | Citation |
|---|---|---|---|---|---|---|
| ALKBH1 | Gastric cancer | mRNA | Upregulation | Not mentioned | Upregulation (OS) | (16) |
| Protein | Downregulation | Tumor size, stage | Not correlated | |||
| ALKBH3 | RCC | Protein | Upregulation | T, M factor, stage | Upregulation (OS) | (17) |
| Bladder cancer (pTa/T1) | Protein | Upregulation | T factor | Upregulation (time to recurrence) | (18) | |
| Pancreas cancer | Protein | Upregulation | T factor, stage | Upregulation (RFS) | (19) | |
| NSCLC | Protein | Upregulation | Not mentioned | Upregulation (RFS: LUAD) | (11) | |
| Hepatocellular cancer | Protein | Upregulation | Differentiation, M factor, stage | Upregulation (OS, RFS) | (20) | |
| ALKBH5 | Pancreas cancer | mRNA | Not mentioned | Not mentioned | Upregulation (OS) | (21) |
| Clear cell RCC | mRNA | Downregulation | M factor | Downregulation (OS, CSS) | (22) | |
| Esophageal cancer | Protein | Upregulation | Not correlated | Upregulation (OS) | (23) | |
| Colon cancer | Protein | Downregulation | M factor, stage | Downregulation (OS, RFS) | (24) | |
| NSCLC | mRNA | Upregulation | Differentiation | Upregulation (OS) | (12) | |
| ALKBH9 | Gastric cancer | Protein | Upregulation | Differentiation, N factor, stage | Upregulation (OS) | (25) |
| Endometrial cancer | Protein | Upregulation | Not correlated | Upregulation (OS, RFS) | (26,27) | |
| Clear cell RCC | mRNA | Downregulation | M factor | Downregulation (OS, CSS) | (22) | |
| ALKBH4 | NSCLC | Protein | Upregulation | Not correlated | Upregulation (OS, RFS: LUAD) | This study |
ALKBH, AlkB homolog; OS, overall survival; RCC, renal cell cancer; RFS, recurrence-free survival; LUAD, lung adenocarcinoma; NSCLC, non-small cell lung cancer; CSS, cancer specific survival.
The expression of ALKBH4 was observed in all four cases of adenocarcinoma in situ. It has been reported that expression of ALKBH3 was also observed in 50% of lesions of carcinoma in situ of prostate cancer (28). ALKBH3 was reported to be a candidate gene associated with the risk of papillary thyroid cancer (29). Furthermore, Li et al. (30) demonstrated that ALKBH9 plays an oncogenic role in acute myeloid leukemia. In contrast, Calvo et al. (31) demonstrated that ALKBH2 and ALKBH3 modestly protect against inflammation-mediated colon carcinogenesis and that their effects are at least additive in the Alkbh2/Alkbh3 double-knockout mice. These facts suggest that the ALKBH family is involved in carcinogenesis or early stage of cancer progression. In fact, in this study, the expression of ALKBH4 was not associated with clinicopathological factors that indicate the progression of cancer.
According to previous reports on NSCLC, the high expression of ALKBH3 protein in LUAD and high expression of ALKBH5 mRNA are associated with a poor prognosis (11,12), which may support our current study results. In order to further validate our study, we reviewed the online database Gene Expression Profiling Interactive Analysis 2 (GEPIA2) (32), which is based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) data, containing the gene expression data and survival information of many cancer types. When analysis was performed in 477 patients with LUAD, the OS rate in patients with higher ALKBH4 mRNA expression (n=239) was comparable to that in patients with lower ALKBH4 mRNA expression (n=238) (Log-rank test; P=0.57, hazard ratio =0.92 for high ALKBH4 expression). This discordant result may be due to the difference between the TCGA database and the current study with regard to the criteria for ALKBH4 expression status, oncological profile, patient demographic characters, and the anticancer treatment.
According to the relevant literature, the ALKBH family in various cancers regulates the cell cycle progression, senescence or apoptosis, and is involved in cancer progression (11,17,19,20,23,33,34). The ALKBH family has also been reported to be involved in cancer progression through the control of angiogenesis (18,19,34). Fujii et al. (35) demonstrated that ALKBH2 is an upstream molecule of the oncoprotein, MUC1, and regulates the cell cycle and epithelial to mesenchymal transition, resulting in the progression of urothelial carcinomas. Chao et al. (36) demonstrated that the m6A demethylase ALKBH5 affects the proliferation and invasion of LUAD cells under intermittent hypoxia by downregulating m6A modification on Forkhead box M1 (FOXM1) mRNA and by promoting the expression of FOXM1. Moreover, Zhu et al. (26) investigated whether estrogen enhances the nuclear localization of ALKBH9 and promotes endometrial cancer cell growth via the mTOR signaling pathway. Furthermore, it is reported that ALKBH2 and ALKBH3 are associated with cisplatin sensitivity (20,37). In addition, Zhou et al. (38) reported that ALKBH9 enhances the chemo-radiotherapy resistance of cervical squamous cell carcinoma both in vitro and in vivo through the regulation of the β-catenin expression by reducing m6A levels in its mRNA transcripts. In contrast, Yang et al. (24) established that the overexpression of ALKBH5 inhibited colon cancer cells invasion in vitro and metastasis in vivo. To date, few reports have examined the role of ALKBH4 in cancer cells. Pilžys et al. (39) reported that the silencing of ALKBH4 markedly decreased HeLa cancer cell survival. We have previously shown that ALKBH4 promotes the progression of lung cancer via E2F1 (14), which plays an important role in cell cycle progression, senescence and apoptosis (40-42). As mentioned above, the ALKBH family is associated with the progression of various cancers by playing a multifunctional role. Therefore, further research is needed to elucidate the mechanism through which ALKBH4 is involved in lung cancer progression.
There are some limitations in this study. First, we retrospectively analyzed relatively small number of patients with resectable lung cancers. Thus, the clinical impact of ALKBH4 in advanced stage lung cancer still remains unknown. Second, in the current study, we did not investigate whether expression status of ALKBH4 was associated with the response to chemotherapy, which could impact on postoperative prognosis. Finally, the current study did not clarify the relationship among ALKBH4 and other ALKBH family.
In conclusion, the expression of ALKBH4 was significantly associated with a poorer prognosis in patients with LUAD, but not in patients with LUSC. The expression of ALKBH4 may be involved in the development and progression of LUAD. Further study on the role of the ALKBH family in the mechanisms of carcinogenesis and therapeutic biomarkers is warranted.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-39/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-39/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-39/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-39/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Committee of Kagoshima University Hospital (registration number 351) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Takano T, Fukui T, Ohe Y, et al. EGFR mutations predict survival benefit from gefitinib in patients with advanced lung adenocarcinoma: a historical comparison of patients treated before and after gefitinib approval in Japan. J Clin Oncol 2008;26:5589-95. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 2011;12:1004-12. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 2002;419:178-82. [Crossref] [PubMed]
- Trewick SC, Henshaw TF, Hausinger RP, et al. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 2002;419:174-8. [Crossref] [PubMed]
- Aas PA, Otterlei M, Falnes PO, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 2003;421:859-63. [Crossref] [PubMed]
- Kurowski MA, Bhagwat AS, Papaj G, et al. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics 2003;4:48. [Crossref] [PubMed]
- Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007;318:1469-72. [Crossref] [PubMed]
- Ougland R, Rognes T, Klungland A, et al. Non-homologous functions of the AlkB homologs. J Mol Cell Biol 2015;7:494-504. [Crossref] [PubMed]
- Tasaki M, Shimada K, Kimura H, et al. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Br J Cancer 2011;104:700-6. [Crossref] [PubMed]
- Zhu Z, Qian Q, Zhao X, et al. N6-methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene 2020;731:144348. [Crossref] [PubMed]
- Li MM, Nilsen A, Shi Y, et al. ALKBH4-dependent demethylation of actin regulates actomyosin dynamics. Nat Commun 2013;4:1832. [Crossref] [PubMed]
- Jingushi K, Aoki M, Ueda K, et al. ALKBH4 promotes tumourigenesis with a poor prognosis in non-small-cell lung cancer. Sci Rep 2021;11:8677. [Crossref] [PubMed]
- Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706-14. [Crossref] [PubMed]
- Li Y, Zheng D, Wang F, et al. Expression of Demethylase Genes, FTO and ALKBH1, Is Associated with Prognosis of Gastric Cancer. Dig Dis Sci 2019;64:1503-13. [Crossref] [PubMed]
- Hotta K, Sho M, Fujimoto K, et al. Clinical significance and therapeutic potential of prostate cancer antigen-1/ALKBH3 in human renal cell carcinoma. Oncol Rep 2015;34:648-54. [Crossref] [PubMed]
- Shimada K, Fujii T, Tsujikawa K, et al. ALKBH3 contributes to survival and angiogenesis of human urothelial carcinoma cells through NADPH oxidase and tweak/Fn14/VEGF signals. Clin Cancer Res 2012;18:5247-55. [Crossref] [PubMed]
- Yamato I, Sho M, Shimada K, et al. PCA-1/ALKBH3 contributes to pancreatic cancer by supporting apoptotic resistance and angiogenesis. Cancer Res 2012;72:4829-39. [Crossref] [PubMed]
- Wang Q, Wang G, Wang Y, et al. Association of AlkB homolog 3 expression with tumor recurrence and unfavorable prognosis in hepatocellular carcinoma. J Gastroenterol Hepatol 2018; Epub ahead of print. [Crossref] [PubMed]
- Cho SH, Ha M, Cho YH, et al. ALKBH5 gene is a novel biomarker that predicts the prognosis of pancreatic cancer: A retrospective multicohort study. Ann Hepatobiliary Pancreat Surg 2018;22:305-9. [Crossref] [PubMed]
- Strick A, von Hagen F, Gundert L, et al. The N6 -methyladenosine (m6 A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int 2020;125:617-24. [Crossref] [PubMed]
- Nagaki Y, Motoyama S, Yamaguchi T, et al. m6 A demethylase ALKBH5 promotes proliferation of esophageal squamous cell carcinoma associated with poor prognosis. Genes Cells 2020;25:547-61. [Crossref] [PubMed]
- Yang P, Wang Q, Liu A, et al. ALKBH5 Holds Prognostic Values and Inhibits the Metastasis of Colon Cancer. Pathol Oncol Res 2020;26:1615-23. [Crossref] [PubMed]
- Xu D, Shao W, Jiang Y, et al. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep 2017;38:2285-92. [Crossref] [PubMed]
- Zhu Y, Shen J, Gao L, et al. Estrogen promotes fat mass and obesity-associated protein nuclear localization and enhances endometrial cancer cell proliferation via the mTOR signaling pathway. Oncol Rep 2016;35:2391-7. [Crossref] [PubMed]
- Zhang Z, Zhou D, Lai Y, et al. Estrogen induces endometrial cancer cell proliferation and invasion by regulating the fat mass and obesity-associated gene via PI3K/AKT and MAPK signaling pathways. Cancer Lett 2012;319:89-97. [Crossref] [PubMed]
- Konishi N, Nakamura M, Ishida E, et al. High expression of a new marker PCA-1 in human prostate carcinoma. Clin Cancer Res 2005;11:5090-7. [Crossref] [PubMed]
- Neta G, Brenner AV, Sturgis EM, et al. Common genetic variants related to genomic integrity and risk of papillary thyroid cancer. Carcinogenesis 2011;32:1231-7. [Crossref] [PubMed]
- Li Z, Weng H, Su R, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 2017;31:127-41. [Crossref] [PubMed]
- Calvo JA, Meira LB, Lee CY, et al. DNA repair is indispensable for survival after acute inflammation. J Clin Invest 2012;122:2680-9. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Gao W, Li L, Xu P, et al. Frequent down-regulation of hABH2 in gastric cancer and its involvement in growth of cancer cells. J Gastroenterol Hepatol 2011;26:577-84. [Crossref] [PubMed]
- Shimada K, Nakamura M, Anai S, et al. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res 2009;69:3157-64. [Crossref] [PubMed]
- Fujii T, Shimada K, Anai S, et al. ALKBH2, a novel AlkB homologue, contributes to human bladder cancer progression by regulating MUC1 expression. Cancer Sci 2013;104:321-7. [Crossref] [PubMed]
- Chao Y, Shang J, Ji W. ALKBH5-m6A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem Biophys Res Commun 2020;521:499-506. [Crossref] [PubMed]
- Wu SS, Xu W, Liu S, et al. Down-regulation of ALKBH2 increases cisplatin sensitivity in H1299 lung cancer cells. Acta Pharmacol Sin 2011;32:393-8. [Crossref] [PubMed]
- Zhou S, Bai ZL, Xia D, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog 2018;57:590-7. [Crossref] [PubMed]
- Pilžys T, Marcinkowski M, Kukwa W, et al. ALKBH overexpression in head and neck cancer: potential target for novel anticancer therapy. Sci Rep 2019;9:13249. [Crossref] [PubMed]
- Wu L, Timmers C, Maiti B, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 2001;414:457-62. [Crossref] [PubMed]
- Park C, Lee I, Kang WK. E2F-1 is a critical modulator of cellular senescence in human cancer. Int J Mol Med 2006;17:715-20. [Crossref] [PubMed]
- Engelmann D, Pützer BM. The dark side of E2F1: in transit beyond apoptosis. Cancer Res 2012;72:571-5. [Crossref] [PubMed]


