
CD13 downregulation mediated by ubenimex inhibits autophagy to overcome 5-FU resistance by disturbing the EMP3/FAK/NF-κB pathway in gastric cancer cells
Introduction
As the fourth most common cancer, gastric cancer (GC) is ranked as the second leading cause of cancer-related death worldwide (1). Patients with early GC are usually asymptomatic, and the rate of early diagnosis of GC is low; most patients (>70%) present with advanced GC (2). Currently, 5-fluorouracil (5-FU)-based chemotherapy is widely used to enhance the quality of life and extend the survival cycle of patients with advanced GC. However, experimental studies in the clinic have revealed that the sensitivity of GC patients to 5-FU is gradually decreasing, making the treatment for GC patients end in failure (3,4). 5-FU resistance has been demonstrated to be a fundamental obstacle in the chemotherapy of GC, and identifying ways to reverse 5-FU resistance in GC has become an important issue to be addressed.
Autophagy is a process by which cells use lysosomal enzymes to degrade harmful substances and maintain a stable intracellular environment (5). The traditional view indicates that autophagy is an adaptive response made by normal cells to tolerate metabolic stress environments that are not conducive to their own survival to maintain the normal physiology of cells (6). However, with the evolving understanding of the molecular mechanisms involved in autophagy, it has been found that autophagy produces a marked effect on the development of drug resistance. To relieve chemotherapy-induced stress, tumour cells activate autophagy to defend against stress, leading to cytoprotective effects called chemoresistance (7). Studies have shown that autophagy mediates tumour escape from anoikis and promotes tumour cell survival; therefore, cell autophagy induces the development of drug resistance in GC, which is closely related to its triggering of apoptosis tolerance (8,9).
CD13, also known as aminopeptidase N (APN), belongs to the class II membrane-bound metalloproteinases. CD13 is overexpressed in liver, stomach, pancreas, colon, prostate and thyroid tumours, and it utilizes the action of proteolytic enzymes to accelerate tumour proliferation, vascularization, metastasis and infiltration (10). A previous study has found that ubenimex, a CD13 inhibitor, synergizes with 5-FU to improve the antitumour activity of 5-FU in liver, kidney and breast cancer cells (11). Ubenimex also targets CD13, promotes apoptosis induced by chemotherapeutic agents and reverses multidrug resistance (MDR) in liver cancer cells (12). However, as the only marketed CD13 inhibitor, ubenimex is only used as an immunomodulatory adjuvant for the treatment of haematological malignancies (13). There are no reports on the reversal of 5-FU resistance mediated by ubenimex.
Initially discovered to undergo hypermethylation-mediated transcriptional silencing in glioma, non-small-cell lung cancer (NSCLC) and oesophageal squamous cell carcinoma (ESCC), epithelial membrane protein 3 (EMP3) has attracted attention as a well-established tumour suppressor (14-16). However, accumulating evidences suggest a tumour-promoting role for EMP3 in the breast, urothelium and hepatocytes (14,17,18). Although there is evidence showing that the mRNA of EMP3 is upregulated in GC-derived cell lines (19), the role of EMP3 in GC malignancy development and its impact on GC treatment are currently unknown.
Focal adhesion kinase (FAK) is a cytoplasmic nonreceptor tyrosine kinase that mediates the adhesion of cells to the extracellular matrix (ECM) and plays a crucial role in regulating the function of basic cells with overexpression in many cancers, including glioblastoma, breast, colorectal, pancreatic, lung and ovarian cancers (20,21). Evidence has indicated that the expression and phosphorylation of FAK play an important role in the initiation and transmission of autophagy signals as well as the induction of apoptosis tolerance in GC cells (22). Blockade of FAK expression and phosphorylation activity inhibits the entry of FAK into the nucleus, which attenuates the transcriptional activity of nuclear factor-κB (NF-κB), thereby downregulating the expression of autophagy proteins, such as Beclin-1, and inhibiting the occurrence of autophagy (23). Among autophagy proteins, Beclin-1 plays a key role in the process of apoptosis resistance induced by autophagic “addiction”. Inhibition of Beclin-1 expression activates caspase-mediated apoptosis in tumour cells, whereas activated caspases induce apoptosis in tumour cells by affecting Beclin-1, thereby interfering with Bcl-2 and Bax protein levels (24,25).
In the present study, we confirmed that ubenimex alleviates GC autophagy to reverse 5-FU resistance by attenuating the expression of CD13. Additionally, we utilized high content screening (HCS) and found that EMP3 is a key molecule in the targeting of CD13 by ubenimex in suppressing GC, and through a series of experiments, we verified that ubenimex may attenuate GC autophagy by affecting the CD13/EMP3/FAK/NF-κB pathway. Collectively, these results indicated that ubenimex is a prospective agent that may attenuate GC autophagy to reverse 5-FU resistance, thus improving GC sensitivity to 5-FU. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-345/rc).
Methods
Reagents and antibodies
Ubenimex was purchased from Shenzhen Main Luck Pharmaceutical, Inc. (Shenzhen, China), and 5-FU was obtained from Xudong Haipu Pharmaceutical Co., Ltd. (Shanghai, China). Rabbit monoclonal antibodies against ANPEP (CD13; Abcam Biotechnology, Cambridge, USA; cat. no. ab108382), LC3B (Abcam; cat. no. ab192890), Bax (ABclonal Technology Co., Ltd., Wuhan, China; cat. no. A19684), p-FAK (Abcam; cat. no. ab81298), Rabbit polyclonal antibodies against EMP3 (Abcam; cat. no. ab236671), PTK2 (Sangon Biotech Co., Ltd., Shanghai, China; KleanAB; cat. no. P102163), NF-κB P65 (Proteintech, Rosemont, IL, USA; cat. no. 10745-1-AP), P62/SQSTM1 (Proteintech; cat. no. 18420-1-AP), Beclin-1 (Proteintech; cat. no. 11306-1-AP), ATG5 (Proteintech; cat. no. 10181-2-AP), Bax (Proteintech; cat. no. 50599-2-Ig), Bcl-2 (ABclonal; cat. no. A0208 and Proteintech; cat. no. 12789-1-AP), GAPDH (ABclonal; cat. no. AC001). All secondary antibodies (cat. no. SA00001-2) were purchased from Proteintech Biosciences.
Establishment of 5-FU-resistant GC cells
The human GC cell line, MKN-45, was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). When MKN-45 cells were in logarithmic growth phase, medium containing 5-FU (2 µg/mL) or oxaliplatin (0.1 µg/mL) was added. The above steps were repeated over 7 months until induction of MDR in cells (designated MKN-45/X) was confirmed. The human GC cell line, SGC-7901, was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). We generated SGC-7901/5-FU cells by exposing the parental cells to increasing concentrations of 5-FU, ranging from 0 to 80 µg/mL, for more than 3 months. The resistance index (RI) was estimated by the following formula: RI = IC50 (resistant cells)/IC50 (parental cells). The morphology and RI of drug-resistant cells are shown in Figure S1A,S1B. Furthermore, CD13 expression was up-regulated in SGC-7901/5-FU cells, compared to the parental GC cells (Figure S1C).
Plasmid construction and gene amplification
Enhanced green fluorescent protein (EGFP)-labeled ANPEP (CD13; GenBank number NM_001150), EMP3 (GenBank number NM_001425) overexpression plasmids and negative control plasmids (empty vector control plasmid; CMV-MCS-3FLAG-IRES-EGFP-SV40-Neomycin) were used in this study. Detailed information is provided in Appendix 1.
Gene chip analysis and HCS of cell function
The Affymetrix ClariomTM S gene chip and the HCS were used to were used to screen the key genes involved in the reversal of resistance to ubenimex. Detailed information is provided in Appendix 1.
Proliferative activity and cell sensitivity assays
The proliferative activity and sensitivity of GC cells to 5-FU were determined by the Cell Counting Kit-8 (CCK-8) assay. Detailed information is provided in Appendix 1.
Western blot analysis
After cells were incubated with different treatments, protein expression levels were determined. The detailed procedure is provided in Appendix 1.
Transmission electron microscopy
The human 5-FU-resistant cell line, SGC-7901/5-FU, was treated with 0.12 mg/mL ubenimex for 24 h. Cells were then fixed with 2.5% glutaraldehyde in PBS buffer for 1.5 h followed by fixation in 1% osmium tetroxide for 1.5 h. Cells were then washed, stained with 3% aqueous uranyl acetate, dehydrated with an increasing concentration gradient of ethanol and embedded in Araldite. Ultrathin sections were observed under a CM-120 electron microscope (Philips, USA).
Annexin V-FITC/PI staining
Annexin V-FITC/PI dual staining was used to determine the effect of ubenimex on apoptosis. Briefly, the indicated cells were treated with different concentrations of 5-FU and ubenimex for 48 h and then harvested. Cells were resuspended in 300 µL of 1× binding buffer, incubated with Annexin V-FITC and PI in the dark for 15 min and then analysed using an EPICS XL flow cytometer (BD Biosciences, USA).
Immunofluorescence analysis
Cell concentration smears were made, and cells were fixed with 4% paraformaldehyde for 20 min. Cells were thoroughly washed with cold phosphate-buffered saline (PBS) (3 min ×3), fixed with 4% paraformaldehyde for 20 min at room temperature and washed with cold PBS (3 min ×3). Goat serum blocking solution (100 µL/sample) was added for 60 min at room temperature followed by incubation with primary antibodies diluted 1:200 in blocking solution at 4 ℃ overnight in the dark. Cells were then washed in cold PBS (3 min ×3) and then incubated 1:200 fluorescent secondary antibody at 100 µL/sample (diluted in PBS) at room temperature in the dark for 1 h. Cells were washed with cold PBS (5 min ×3) and then incubated with 4',6-diamidino-2-phenylindole (DAPI) for 15 min. Cells were washed with cold PBS (3 min ×3) and then incubated with an anti-fluorescent quencher.
Statistical analysis
GraphPad Prism 7.0 software (La Jolla, CA, United States) was utilized for statistical analysis (GraphPad Prism, RRID:SCR_002798). Each group of independent experiments was performed at least three times. All data are presented as the means ± standard deviation (SD) and were evaluated by one-way analysis of variance. P<0.05 indicated statistical significance. Statistically significant P values are presented as *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
Results
Ubenimex reverses the drug resistance of SGC-7901/5-FU cells
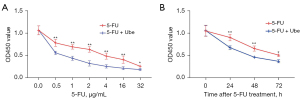
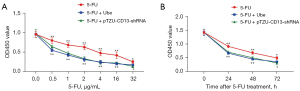
Ubenimex acts as a CD13 inhibitor to induce immune activation by stimulating CD16+CD56+ NK cells and CD3+CD4+ T lymphocytes. Ubenimex can be used as an immunopotentiator for the treatment of leukaemia and multiple myeloma. However, there are currently few studies on the use of ubenimex for the treatment of GC or reversal of drug resistance in GC-resistant cells. We next investigated the effects of ubenimex on 5-FU sensitivity. SGC-7901/5-FU cells were incubated with various concentrations of 5-FU (0.5, 1, 2, 4, 16 and 32 µg/mL) for 48 h in the absence or presence of ubenimex (0.12 mg/mL) or were incubated with 5-FU (12 µg/mL) for 0–72 hours in the absence or presence of ubenimex (0.12 mg/mL). The CCK-8 results showed that ubenimex significantly increased the sensitivity of SGC-7901/5-FU cells to 5-FU in a time- and dose-dependent manner (Figure 1A,1B). Therefore, these findings suggested that ubenimex may reverse the drug resistance of acquired drug-resistant GC cells.
Ubenimex inhibits autophagic death in SGC-7901/5-FU cells
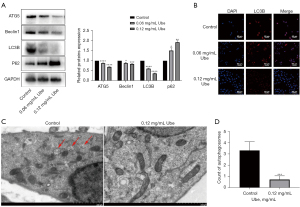
Previous experimental evidence has demonstrated that ubenimex inhibits autophagic cell death (26). In the present study, western blot analyses, cellular immunofluorescence and transmission electron microscopy were used to examine the autophagy level in SGC-7901/5-FU cells after 48 h of exposure to ubenimex following pretreatment with 5-FU. With increasing doses of ubenimex, LC3B, Beclin-1 and ATG5 levels decreased in SGC-7901/5-FU cells, whereas the levels of P62 showed a tendency to increase (Figure 2A). These findings indicated that the increased dose of ubenimex plays a key role in inhibiting a higher level of autophagic cell death in GC-resistant cells. LC3B attachment to the membrane is widely regarded as a sign of autophagosome formation (27). Hence, SGC-7901/5-FU cells were treated the same as described above and further examined LC3B expression levels by immunofluorescence. We found that the results were consistent with previous conclusions (Figure 2B). In addition, transmission electron microscopy was used to observe a decrease in the number of autophagosomes in ubenimex-treated cells (P<0.001; Figure 2C,2D), which suggested that autophagy was reduced after ubenimex treatment. Together, these data indicated a critical role of autophagy in ubenimex-regulated 5-FU sensitivity in GC-resistant cells.
Ubenimex induces apoptosis mediated by chemotherapeutic drugs in SGC-7901/5-FU cells
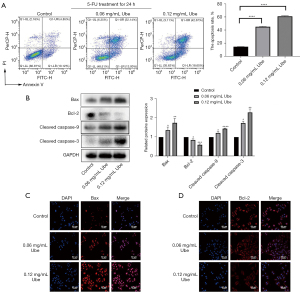
Cells were treated with Annexin V-FITC/PI double staining to quantify the level of apoptosis. After pretreatment with 5-FU, ubenimex induced apoptosis of SGC-7901/5-FU cells in a dose-dependent manner. After 48 h of exposure to ubenimex, the increased dose (0.12 mg/mL) of ubenimex resulted in a more than fourfold increase in the proportion of total apoptotic cells compared to control cells, and the majority of SGC-7901/5-FU cells were characterized as a late apoptotic population (Figure 3A). In addition, the expression of major apoptotic proteins namely, cleaved caspase-3, cleaved caspase-9, Bax and Bcl-2, was further examined by western blot analysis to explore the effect of ubenimex on apoptosis. As the concentration of ubenimex increased, the expression of the Bcl-2 antiapoptotic protein decreased, while the levels of cleaved caspase-3, cleaved caspase-9 and Bax were significantly upregulated (Figure 3B), which further corroborated that ubenimex treatment induced apoptotic effects. The immunofluorescence results also showed that the expression of Bax was elevated and that the expression of Bcl-2 was decreased (Figure 3C,3D). These results revealed that ubenimex plays a key role in inducing apoptosis by 5-FU in GC-resistant cells.
Screening of target genes involved in the reversal of drug resistance by ubenimex
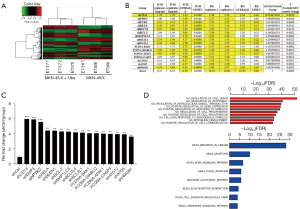
To obtain the target gene related to GC drug resistance, we used microarray analysis to identify the gene expression profile of MKN-45/X cells after ubenimex treatment. Ubenimex caused the downregulation of 264 genes, including CD13, and the upregulation of 228 genes (Figure 4A). We evaluated the changes in RI values of MKN-45/X cells after overexpression or silencing of relevant genes by HCS experiments. We screened the top 16 genes, including CD13, that were related to the reversal of the resistance effect of ubenimex in GC (Figure 4B,4C). Through GO function and KEGG pathway enrichment analyses, we found that the above genes were all enriched in the NF-κB pathway and involved in the regulation of cell autophagy and apoptosis (Figure 4D). After silencing the CD13 gene, we utilized HCS to observe its effect on the expression of other related genes and ranked them by the magnitude of the effect. We found that EMP3 may be a key molecule in the targeting of CD13 by ubenimex and reversing drug resistance in GC (Figure S2A). Similarly, we investigated the impact of silencing EMP3 on related genes and found that the mRNA level of PTK2 (the encoding gene of FAK) was downregulated (Figure S2B), indicating the presence of a potential CD13/EMP3/FAK/NF-κB regulatory axis.
Ubenimex inhibits CD13/EMP3/FAK/NF-κB signalling pathway activity in SGC-7901/5-FU cells
To validate the potential CD13/EMP3/FAK/NF-κB regulatory axis identified by microarray analysis, GO functional analysis and KEGG pathway enrichment analysis, we explored the relationship between CD13 and EMP3 by western blot analysis after overexpression of these proteins. Figure 5A shows that the expression levels of EMP3, FAK and NF-κB were increased after CD13 overexpression. EMP3 overexpression had no significant effect on the expression of CD13 protein, but EMP3 overexpression significantly increased the expression levels of FAK and NF-κB (Figure 5B). These results suggested that CD13 has a one-way regulatory effect on EMP3 expression. Furthermore, western blot assays confirmed that the expression of CD13, EMP3, FAK and NF-κB was downregulated in GC-resistant cells treated with ubenimex at different concentrations (Figure 5C). Interestingly, CD13 overexpression reversed the inhibitory effect of ubenimex on CD13, EMP3, FAK and NF-κB. However, the inhibitory effect of ubenimex on CD13 was not significantly affected after EMP3 overexpression, but EMP3 overexpression reversed the inhibitory effect of ubenimex on EMP3, FAK and NF-κB (Figure 5D,5E). Western blot analysis further confirmed that ubenimex affects the expression of downstream pathway proteins by targeting CD13/EMP3. Moreover, CD13 silencing and ubenimex treatment both attenuated the expression of EMP3, FAK and NF-κB, but the effect mediated by ubenimex was suppressed after CD13 overexpression (Figure 5F). Taken together, these findings showed that ubenimex inhibits EMP3/FAK/NF-κB signalling pathway activity by targeting CD13 in SGC-7901/5-FU cells.
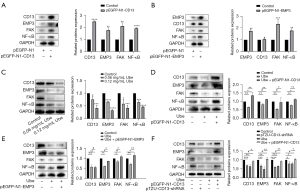
Ubenimex suppresses autophagy to promote 5-FU-induced apoptosis by inhibiting the activation of the CD13/EMP3/FAK/NF-κB pathway
Accumulating evidence suggests that the expression and phosphorylation of FAK play important roles in the initiation and transmission of autophagy signals as well as the induction of apoptosis tolerance in GC cells (22). Immunofluorescence staining of SGC-7901/5-FU cells was performed to test whether ubenimex affects the phosphorylation of FAK by targeting CD13. In both the ubenimex-treated and CD13-deficient groups, the phosphorylation of FAK was significantly downregulated. Notably, the inhibition by ubenimex was attenuated upon CD13 overexpression (Figure 6A). We next investigated whether ubenimex attenuates autophagy in GC-resistant cells by modulating the CD13/EMP3/FAK/NF-κB pathway. Figure 6B shows that there was a significant increase in SQSTM1 expression but a significant reduction in LC3B, Beclin-1 and ATG5 expression in the ubenimex-treated 5-FU-resistant GC cells compared to the control cells, which indicated the downregulation of autophagy. As expected, the results in the CD13-deficient group were consistent with this finding. More importantly, CD13 overexpression reversed the inhibition of autophagy by ubenimex. We completed further validation by detecting LC3B by immunofluorescence (Figure 6C). Together, these results suggested a critical role of autophagy in ubenimex-increased 5-FU sensitivity in GC-resistant cells. It is well known that dysregulated expression of the Bcl-2 family and subsequent inactivation of caspase-3 or caspase-9 are fundamental mechanisms for apoptosis resistance in GC cells (28,29). In the present study, ubenimex increased the expression of Bax, but it downregulated the expression of Bcl-2. Similar to the CD13-deficient group, ubenimex also significantly elevated the expression of the cleaved forms of PARP, caspase-3, and caspase-9, but this effect was attenuated by CD13 overexpression (Figure 6D). In addition, we used Annexin V/PI staining to assess the effects of CD13 silencing, CD13 overexpression and ubenimex treatment on cell apoptosis rates after 5-FU treatment. Ubenimex enhanced the 5-FU-induced apoptosis in GC-resistant cells, while the effect was attenuated by upregulating CD13, which indicated a role for CD13 in 5-FU-induced apoptosis (Figure 6E). Collectively, these results suggested that ubenimex inhibited autophagy to promote 5-FU induced apoptosis, in which the activity of the CD13/EMP3/FAK/NF-κB pathway is alleviated.

Ubenimex renders GC-resistant cells sensitive to 5-FU by alleviating the activity of the CD13/EMP3/FAK/NF-κB pathway
To determine whether ubenimex reverses drug resistance by targeting CD13 and regulating the EMP3/FAK/NF-κB pathway to enhance cellular sensitivity to 5-FU, we downregulated CD13 expression in SGC-7901/5-FU cells. Control cells, ubenimex-treated cells and CD13-deficient cells were subjected to the CCK-8 cell viability assay after pretreatment with different concentrations of 5-FU. Compared to control cells, GC-resistant cells showed increased 5-FU sensitivity after knockdown of CD13 or treated with ubenimex (P<0.05; Figure 7A). However, there was no significant difference between the CD13-deficient and ubenimex-treated groups. In addition, the 5-FU pretreatment group was also tested in the same manner to corroborate whether CD13 knockdown has the same effect on ubenimex-enhanced 5-FU sensitivity and correlates with treatment time. All three groups were treated with 5-FU (12 µg/mL) for 0–72 hours (P<0.05; Figure 7B), and the results indicated that ubenimex enhanced the chemosensitivity of SGC7901/5-FU cells and that CD13 is an essential key target, which was consistent with previous findings.
Discussion
GC is identified as the second leading cause of cancer incidence and mortality, resulting in a serious threat to public health (30). 5-FU is one of the most widely used chemotherapeutic agents for the treatment of GC, which inhibits tumour proliferation and DNA replication by inhibiting thymidylate synthase to synthesize thymine, ultimately inducing apoptosis (31). However, the response rate to 5-FU chemotherapy in patients with advanced GC is less than 32% not satisfied (32). The development of drug resistance in cancer is a complex process in which multiple factors participate, including drug resistance-associated proteins, dysfunction of DNA damage repair, cell autophagy, decreased drug accumulation, metabolic detoxification, alterations in drug targets and signal transduction molecules. This is the first study to suggest that role of CD13-induced autophagy in the 5-FU resistance of GC cells is to protect tumour cells against apoptosis and that autophagy induced by 5-FU is a protective mechanism conducive to cell survival. Consistently, we also found that ubenimex, a CD13 inhibitor, reverses 5-FU resistance and significantly increases the sensitivity of SGC-7901/5-FU cells to 5-FU in a dose- and time-dependent manner.
As recognized, FAK expression and phosphorylation activity are important for the initiation and transmission of autophagy signals as well as the induction of apoptosis tolerance in GC cells, which can affect the expression of NF-κB, whereas the NF-κB, p65 and E2F transcription factors upregulate BECN1 gene expression and increase the level of autophagy (23). As one of the key molecules in autophagosome nucleation, Beclin-1 is an important target in the regulation of autophagy. ATG5 is one of the critical regulators of autophagic cell death and is a protective molecule in tumour cells during the course of chemotherapy. Upregulated expression of ATG5 in GC tissues is associated with chemoresistance in GC and has an important protective role in autophagy (33). p62, another important autophagy-related protein, is an autophagy substrate that is degraded in autophagosomes, and it is considered an indicator of autophagic flux (34). It has been shown that high p62 expression is closely related to poor prognosis in oral squamous cell carcinoma and triple-negative breast cancer but not in colon and breast cancers (35).
Apoptosis refers to the process of genetically controlled, autonomous and orderly death of multicellular organisms under physiological or pathological changes in which caspase-dependent apoptotic pathway is well documented. In particular, the caspase family-mediated proteasome cleavage cascade, ultimately leading to apoptosis. Evidence has shown that the signals of each apoptotic pathway eventually converge to caspase-3 to execute apoptosis, and the activation of caspase-3 may destroy its substrate, PARP, leading to the loss of DNA repair function and cell apoptosis (36). The Bcl-2 gene family is one of the main regulators of the mitochondrial pathway, and its family members are divided into two main categories as follows: anti-apoptotic proteins (such as Bcl-2) and pro-apoptotic proteins (such as Bax). Both Bcl-2 and Bax interact with each other on mitochondria, and their regulation of apoptosis is not only dependent on the expression level of the gene but also related to the ratio between the two.
Apoptosis and autophagy are two important cellular biological behaviours that maintain the stability of the internal environment, and they exist in the physiological process of normal cells and are also an important defence mechanism for cells under injury. However, once a tumour is formed or when the external environment disturbs cellular regulation, autophagy provides abundant nutrition and energy for cancer cells to promote cell survival. Recent evidence has suggested that autophagy plays a dual role in promoting or inhibiting apoptosis. Autophagy mainly plays an anti-apoptotic role by degrading apoptosis-related proteins, which contribute to the occurrence of acquired drug resistance. As a caspase substrate, Beclin-1 is an important molecule in autophagy because it can be cleaved by a caspase. After cleavage, Beclin-1 loses autophagy function but promotes cell apoptosis (37). As an important tumour suppressor protein, Beclin-1 not only interacts with the apoptosis-related genes, Bcl-2 and caspase-9, but also participates in the formation of the autophagy bilayer membrane and jointly participates in the regulation of apoptosis and autophagy (38).
Here, the genes related to the ubenimex-induced reversal of GC resistance were screened using microarray analysis and HCS. EMP3 and FAK were identified to be the key molecules involved in the ubenimex-induced reversal of GC resistance by targeting CD13, and a positive correlation between FAK and EMP3 expression was also found in GC patient tissues. However, studies on the relationship between EMP3 and 5-FU-resistant GC cells as well as the key mechanism by which EMP3 regulates FAK activity are limited. Previous evidence has demonstrated that the aberrant expression of EMP3 in GC cells is partially attributed to hypermethylation impairment of its silencing mechanism (39). Additionally, CD13 may be the initiator of harm in this process by further regulating NF-κB expression, nuclear import and its transcriptional activity by affecting FAK expression and phosphorylation activity. The possibility of a CD13/EMP3/FAK/NF-κB regulatory axis was also further supported by western blot analysis in the present study using CD13 and EMP3 overexpression and silencing vectors, which showed that ubenimex increased its inhibitory effect on pathway activity in a dose-dependent manner. Therefore, these findings suggested that ubenimex enhances 5-FU sensitivity and thus reverses drug resistance by inhibiting the activation of the EMP3/FAK/NF-κB pathway in GC cells by targeting CD13. To the best of our knowledge, this is the first study to investigate the underlying molecular mechanisms of ubenimex-induced chemosensitivity of GC cells to 5-FU via CD13/EMP3/FAK/NF-κB-mediated transcriptional activation. Furthermore, the changes in the ATG5, Beclin-1, LC3B and p62 autophagy-related proteins after treatment with ubenimex, CD13 overexpression and CD13 silencing were determined using western blot and immunofluorescence analyses. Finally, the number of autophagosomes was examined by transmission electron microscopy. These results demonstrated that ubenimex suppresses the occurrence of autophagy in GC drug-resistant cells by targeting CD13.
Consistently, the apoptosis of GC drug-resistant cells was detected by Annexin V/PI double staining, and flow cytometry analysis showed that SGC-7901/5-FU cells treated with ubenimex had increased apoptosis compared to control cells, and the effect of apoptosis increased with the concentration of ubenimex. Western blot and immunofluorescence analyses also showed increased expression of Bax, cleaved PARP, cleaved caspase-3 and cleaved caspase-9 as well as decreased expression of Bcl-2, indicating a mixed phenotype of autophagic cell death and apoptosis by targeting CD13 in response to treatment with ubenimex in SGC-7901/5-FU cells. Importantly, we found that ubenimex significantly attenuated the autophagic properties of 5-FU in GC-resistant cells. Therefore, we hypothesized that ubenimex may be a reliable autophagy inhibitor and promote 5-FU-induced apoptosis.
The present study showed that FAK expression and phosphorylation play a key role in regulating autophagy and apoptosis. In the present study, immunofluorescence detection of p-FAK expression indicated that FAK phosphorylation was significantly decreased after ubenimex treatment and CD13 silencing, but these effects were reversed after CD13 overexpression. These findings indirectly demonstrated that ubenimex inhibits autophagy by regulating the EMP3/FAK/NF-κB pathway by targeting CD13 to promote chemotherapeutic drug-induced apoptosis and reverse chemoresistance. However, the detailed mechanism by which ubenimex induces apoptosis by inhibiting autophagy needs to be further studied.
Conclusions
Our findings demonstrated that autophagy may be suppressed, at least in part, by interfering with the CD13/EMP3/FAK/NF-κB axis, which attributes to the poor response to 5-FU-based chemotherapy in patients. Overall, the present study suggested that the combination of 5-FU and ubenimex may represent an innovative therapeutic strategy for GC patients and provide attractive insights regarding the combined use of Ubenimex as a GC chemotherapeutic intervention.
Acknowledgments
Funding: This work was supported by Shandong Provincial Natural Science Foundation of China (No. ZR2020QH362).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-345/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-345/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-345/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Machlowska J, Baj J, Sitarz M, et al. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci 2020;21:4012. [Crossref] [PubMed]
- Tan Z. Recent Advances in the Surgical Treatment of Advanced Gastric Cancer: A Review. Med Sci Monit 2019;25:3537-41. [Crossref] [PubMed]
- Xuan Y, Hur H, Ham IH, et al. Dichloroacetate attenuates hypoxia-induced resistance to 5-fluorouracil in gastric cancer through the regulation of glucose metabolism. Exp Cell Res 2014;321:219-30. [Crossref] [PubMed]
- Khakbaz P, Panahizadeh R, Vatankhah MA, et al. Allicin Reduces 5-fluorouracil-resistance in Gastric Cancer Cells through Modulating MDR1, DKK1, and WNT5A Expression. Drug Res (Stuttg) 2021;71:448-54. [Crossref] [PubMed]
- Marinković M, Šprung M, Buljubašić M, et al. Autophagy Modulation in Cancer: Current Knowledge on Action and Therapy. Oxid Med Cell Longev 2018;2018:8023821. [Crossref] [PubMed]
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 2009;335:1-32. [Crossref] [PubMed]
- Qin L, Xu T, Xia L, et al. Chloroquine enhances the efficacy of cisplatin by suppressing autophagy in human adrenocortical carcinoma treatment. Drug Des Devel Ther 2016;10:1035-45. [PubMed]
- D'Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int 2019;43:582-92. [Crossref] [PubMed]
- Xu J, Zhang G, Tong Y, et al. Corilagin induces apoptosis, autophagy and ROS generation in gastric cancer cells in vitro. Int J Mol Med 2019;43:967-79. [PubMed]
- Guzman-Rojas L, Rangel R, Salameh A, et al. Cooperative effects of aminopeptidase N (CD13) expressed by nonmalignant and cancer cells within the tumor microenvironment. Proc Natl Acad Sci U S A 2012;109:1637-42. [Crossref] [PubMed]
- Wang X, Jing F, Zhu H, et al. Activity screening and structure-activity relationship of the hit compounds targeting APN/CD13. Fundam Clin Pharmacol 2011;25:217-28. [Crossref] [PubMed]
- Guo Q, Sui ZG, Xu W, et al. Ubenimex suppresses Pim-3 kinase expression by targeting CD13 to reverse MDR in HCC cells. Oncotarget 2017;8:72652-65. [Crossref] [PubMed]
- Yamazaki T, Sugiyama K, Ichihara K. Effect of ubenimex on the immune system of patients with hematological malignancies. Biomed Pharmacother 1991;45:105-12. [Crossref] [PubMed]
- Hsieh YH, Hsieh SC, Lee CH, et al. Targeting EMP3 suppresses proliferation and invasion of hepatocellular carcinoma cells through inactivation of PI3K/Akt pathway. Oncotarget 2015;6:34859-74. [Crossref] [PubMed]
- Jiang Z, Zhou W, Li XG, et al. The methylation analysis of EMP3 and PCDH-gamma-A11 gene in human glioma. Zhonghua Wai Ke Za Zhi 2010;48:300-4. [PubMed]
- Xue Q, Zhou Y, Wan C, et al. Epithelial membrane protein 3 is frequently shown as promoter methylation and functions as a tumor suppressor gene in non-small cell lung cancer. Exp Mol Pathol 2013;95:313-8. [Crossref] [PubMed]
- Zhou W, Jiang Z, Li X, et al. EMP3 overexpression in primary breast carcinomas is not associated with epigenetic aberrations. J Korean Med Sci 2009;24:97-103. [Crossref] [PubMed]
- Wang YW, Li WM, Wu WJ, et al. Potential significance of EMP3 in patients with upper urinary tract urothelial carcinoma: crosstalk with ErbB2-PI3K-Akt pathway. J Urol 2014;192:242-51. [Crossref] [PubMed]
- Mikata R, Yokosuka O, Fukai K, et al. Analysis of genes upregulated by the demethylating agent 5-aza-2'-deoxycytidine in gastric cancer cell lines. Int J Cancer 2006;119:1616-22. [Crossref] [PubMed]
- Tai YL, Chen LC, Shen TL. Emerging roles of focal adhesion kinase in cancer. Biomed Res Int 2015;2015:690690. [Crossref] [PubMed]
- Romaniuk-Drapała A, Totoń E, Konieczna N, et al. hTERT Downregulation Attenuates Resistance to DOX, Impairs FAK-Mediated Adhesion, and Leads to Autophagy Induction in Breast Cancer Cells. Cells 2021;10:867. [Crossref] [PubMed]
- Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008;181:497-510. [Crossref] [PubMed]
- Copetti T, Bertoli C, Dalla E, et al. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol 2009;29:2594-608. [Crossref] [PubMed]
- Kasprowska-Liśkiewicz D. The cell on the edge of life and death: Crosstalk between autophagy and apoptosis. Postepy Hig Med Dosw (Online) 2017;71:825-41. [Crossref] [PubMed]
- Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell 2012;3:17-27. [Crossref] [PubMed]
- Han L, Zhao Q, Liang X, et al. Ubenimex enhances Brd4 inhibition by suppressing HEXIM1 autophagic degradation and suppressing the Akt pathway in glioma cells. Oncotarget 2017;8:45643-55. [Crossref] [PubMed]
- Ladoire S, Chaba K, Martins I, et al. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy 2012;8:1175-84. [Crossref] [PubMed]
- Florou D, Patsis C, Ardavanis A, et al. Effect of doxorubicin, oxaliplatin, and methotrexate administration on the transcriptional activity of BCL-2 family gene members in stomach cancer cells. Cancer Biol Ther 2013;14:587-96. [Crossref] [PubMed]
- Hassan M, Watari H, AbuAlmaaty A, et al. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int 2014;2014:150845. [Crossref] [PubMed]
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700-13. [Crossref] [PubMed]
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003;3:330-8. [Crossref] [PubMed]
- Kang BW, Kim JG, Kwon OK, et al. Non-platinum-based chemotherapy for treatment of advanced gastric cancer: 5-fluorouracil, taxanes, and irinotecan. World J Gastroenterol 2014;20:5396-402. [Crossref] [PubMed]
- Ge J, Chen Z, Huang J, et al. Upregulation of autophagy-related gene-5 (ATG-5) is associated with chemoresistance in human gastric cancer. PLoS One 2014;9:e110293. [Crossref] [PubMed]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010;140:313-26. [Crossref] [PubMed]
- Luo RZ, Yuan ZY, Li M, et al. Accumulation of p62 is associated with poor prognosis in patients with triple-negative breast cancer. Onco Targets Ther 2013;6:883-8. [PubMed]
- Choudhary GS, Al-Harbi S, Almasan A. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods Mol Biol 2015;1219:1-9. [Crossref] [PubMed]
- Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010;29:1717-9. [Crossref] [PubMed]
- Huang X, Qi Q, Hua X, et al. Beclin 1, an autophagy-related gene, augments apoptosis in U87 glioblastoma cells. Oncol Rep 2014;31:1761-7. [Crossref] [PubMed]
- Guo Q, Jing FJ, Xu W, et al. Ubenimex induces autophagy inhibition and EMT suppression to overcome cisplatin resistance in GC cells by perturbing the CD13/EMP3/PI3K/AKT/NF-κB axis. Aging (Albany NY) 2019;12:80-105. [Crossref] [PubMed]


