
E2F1 as a potential prognostic and therapeutic biomarker by affecting tumor development and immune microenvironment in hepatocellular carcinoma
Introduction
Primary liver cancer is perceived as one of the most lethal cancers, which is ranked the seventh and the second of new cases and deaths respectively among all malignancies worldwide (1). Liver hepatocellular carcinoma (LIHC) is the major subtype of primary liver cancer, accounting for approximately 75% (2). In recent years, novel treatment strategies including immune-checkpoint inhibitors, tyrosine kinase inhibitors and monoclonal antibodies have been applied to the treatment for LIHC and have achieved great progress (3); however, the outcome remains poor with a 5-year survival only standing at about 20% in the United States (4).
Since 50–60% LIHC patients need to be exposed to systemic therapies throughout their entire treatment (3), it is particularly necessary to identify more candidate anti-LIHC targets for effective diagnosis, prognosis, and treatment (5). Nevertheless, approximately 25% of LIHC exist with potentially actionable mutations, which are yet to be translated into the clinical practice (6). Therefore, identifying vital biomarkers and exploring their latently clinical significance could facilitate the development of novel precision treatments.
E2F family of transcription factors plays a crucial role in cell differentiation, response to DNA damage, and cell life cycle (7). A variety of E2F isoforms have been characterized and can be found in most cell types (8). It has been demonstrated that E2F family genes are overexpressed and associated with the development and prognosis in several types of cancers including LIHC (9-13).
Despite that E2F1 is the classic and the most studied E2F member, the role of E2F1 in LIHC is controversial. Some studies reported that E2F1 could promote the proliferation of LIHC by activating ERK/mTOR, B-Myb, BRCA1, Stathmin 1 (14-17) or could inhibit c-Myc-driven apoptosis via activating PIK3CA/Akt/mTOR and COX-2 pathway (18). Meanwhile, several studies indicated that E2F1 exhibited tumor-suppressing activity in LIHC. For instance, TFDP3/E2F1 pathway could promote LIHC cell apoptosis by upregulating HIF-2α, and HIF-2α overexpression was associated with favorable overall survival (OS) of LIHC patients (19). Thus, more studies are needed to further verify the exact role of E2F1 in LIHC (13).
Bioinformatics is an effective way to integrate and process vast life sciences data (20). Thus, this study adopted bioinformatics to merge disparate data sources to explore the possible roles of E2F1 in LIHC. Hub genes in LIHC were identified followed by exploring the expression level and prognostic value of E2F1. Moreover, possible regulating mechanisms including promotor methylation, mutation, copy number alterations, and competing endogenous RNAs (ceRNAs) of E2F1 were investigated. In addition, the association between E2F1 and tumor immune microenvironment was examined using single cell sequencing data. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-218/rc).
Methods
Identification of overexpressed genes in LIHC
Two Gene Expression Omnibus (GEO) series, named GSE46408 (21) and GSE54238 (22,23), containing LIHC and normal liver tissue gene expression profiles were selected from GEO (RRID:SCR_004584) (24). Six LIHC tissue samples and 6 paired normal liver parenchyma samples of GSE46408, as well as 13 advanced LIHC tissue samples and 10 normal liver samples of GSE54238 were respectively compared by GEO2R (25) to screen differentially expressed genes (DEGs). Significantly overexpressed DEGs were chosen by the criteria of adjusted P value <0.05 and log2FC ≥1.5 (FC: fold change). Some genes displayed multiple log2FC values in one GEO series due to the use of different probes. Those genes that displayed conflicting log2FC values, some of which >0 (representing overexpression in LIHC) and the others <0 (indicating under-expression in LIHC), were excluded. The shared overexpressed DEGs in 2 GEO series were chosen for further study.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
The biological process, cellular component, molecular function annotations of GO, as well as KEGG pathway enrichment were adopted to analyze the functions of the shared overexpressed DEGs using WebGestalt 2019 (RRID:SCR_006786) (26). Parameters are as following: organism: homo sapiens, method: over-representation analysis, reference set: genome protein coding. The results were visualized through R programming.
PPI network construction and hub genes selection
STRING database Version 11.5 (RRID:SCR_005223) (27) was applied to construct protein-protein interaction (PPI) network of the shared overexpressed DEGs with the following parameters: network type: full STRING network, active interaction sources: all, interaction score: at least 0.700 (high confidence). The STRING analyzing result including nodes (genes) and experimentally determined interaction parameters was input into Cytoscape software (RRID:SCR_003032) (28) with CytoHubba plugin (RRID:SCR_017677). Calculating via betweenness methods, hub genes were selected with the interaction score >160.
Hub genes validation
Next, to verify the expression of selected hub genes, the difference of mRNA expressions of each hub gene in LIHC and normal liver tissue were compared via Oncomine (RRID:SCR_007834) (29) and GEPIA2 (RRID:SCR_018294) (30) respectively. Besides, immunohistochemical staining images in Human Protein Atlas (HPA, RRID:SCR_006710) (31-33) were adopted to illustrate the protein expression of each hub gene in LIHC and normal liver tissue.
Analysis of E2F1 mRNA and protein expression in various cancers including LIHC
The mRNA expression levels of E2F1 in various cancers including LIHC and paired normal tissues were compared by Oncomine, GEPIA2, and TIMER2.0 (RRID:SCR_018737) (34). In Oncomine, the thresholds were set as: P value of 1E-4, fold change of 2, gene rank of top 10%, and data type of all. TIMER2.0 used Wilcoxon test to count E2F1 expression difference between cancer and the corresponding normal tissue. Besides, to verify E2F1 expression level in LIHC, TCGA data containing E2F1 mRNA expression level and clinicopathological features of LIHC were synthesized and analyzed through UALCAN web (RRID:SCR_015827) (35), and were graphically illustrated with Graphpad Prism Version 9.2.0 (RRID:SCR_002798). Moreover, the E2F1 protein expressions in different kinds of cancers including LIHC were traced from HPA, which present the percentage of patients with high or medium immunohistochemical staining using 3 different E2F1 antibodies (HPA008003, CAB000329, CAB019308) respectively.
Exploring the relationship between E2F1 expression and survival in LIHC
The prognostic value of E2F1 in LIHC was probed using Kaplan-Meier plotter (RRID:SCR_018753) (36), which integrated gene expression and clinical data simultaneously from TCGA database (RRID:SCR_003193), GEO database, and European Genome-Phenome Archive (EGA, RRID: SCR_004944) database. The OS, disease-specific survival (DSS), progression-free survival (PFS), and relapse-free survival (RFS) in each clinicopathological characteristic cohort were analyzed according to E2F1 expression level by the following parameters: cut-off: median, RNAseq ID: 1869, and default settings for all other parameters.
Moreover, the clinical data and E2F1 expression data were retrieved from TCGA database. Multivariate Cox regression analysis was carried out by SPSS Version 26 (RRID:SCR_019096) to analyze whether E2F1 expression level could serve as an independent prognostic factor for LIHC. High and low E2F1 expression subgroups were categorized based on the median expression level of E2F1.
Investigating E2F1 promotor methylation level in LIHC
E2F1 promotor methylation levels in LIHC and normal liver tissue were investigated from TCGA database via UALCAN web. Results were graphically presented through Graphpad Prism Version 9.2.0. In addition, 373 LIHC samples containing data of mRNA Expression z-Scores (RNA Seq V2 RSEM) and Methylation (HM450) beta-value simultaneously were obtained from TCGA Firehose Legacy Cohort via cBioPortal (RRID:SCR_014555) (37,38). Methylation (HM450) beta-value was normalized into 0 to 1. Then simple linear regression was performed by SPSS Version 26 to calculate the correlation between promotor methylation and mRNA expression and visualized by Graphpad Prism Version 9.2.0.
Analysis of mutations and copy number alterations (CNAs) of E2F1 in LIHC
916 LIHC samples of 914 patients in 3 studies [TCGA, Firehose Legacy; AMC, Hepatology 2014 (39); INSERM, Nat Genet 2015 (40)] were adopted to analyze the frequency and types of E2F1 mutation via cBioPortal. Following that, 673 samples of 671 patients in 2 LIHC studies (TCGA, Firehose Legacy; AMC, Hepatology 2014) were used to acquire the frequency of copy-number alterations. Meanwhile, E2F1 mRNA expression levels in different copy number groups of the TCGA Firehose Legacy Study were compared with One-way ANOVA using SPSS Version 26. Moreover, 349 samples of the TCGA Firehose Legacy Study containing mRNA expression data and copy-number alterations data simultaneously were retrieved from cBioPortal, the correlation of the relative linear copy-number values and the mRNA expression z-scores (RNA Seq V2 RSEM) was analyzed by simple linear regression via SPSS Version 26 and exhibited via Graphpad Prism Version 9.2.0.
Exploring possible ceRNAs of E2F1
According to the mechanism of ceRNAs (41), it is assumed that long non-coding RNAs (lncRNAs) as ceRNAs (ce-lncRNAs) could negatively affect the expression of certain microRNAs (miRNAs) which then negatively affect E2F1 levels. Thus, ce-lncRNA levels should be raised and certain miRNA levels should be lowered. ENCORI (RRID:SCR_016303) (42) was employed to explore possible lncRNAs as ceRNAs (ce-lncRNAs) with the following parameters: genome of human, assembly of hg19, miRNA number ≥2, P value ≤0.01, FDR ≤0.01, and with at least one cancer type data. Following that, only those ce-lncRNAs showing positive coefficient with E2F1 expression level (P value ≤0.01 and FDR ≤0.01) in LIHC were supposed to be ce-lncRNAs. Next, miRNAs which could be regulated by the aforementioned ce-lncRNAs were retrieved from ENCORI. The expression of those miRNAs was investigated through miRCancer database (http://mircancer.ecu.edu/) (43), only those downregulated miRNAs in LIHC were selected for further study. Then, the relationship between those miRNAs’ expression level and OS was investigated using Kaplan-Meier plotter, those miRNAs showing favorable OS for LIHC patients were included into ceRNAs network.
Exploring the association between E2F1 and tumor immune microenvironment via single cell sequencing data
Four human LIHC datasets (LIHC_GSE125449_aPDL1aCTLA4, LIHC_GSE140228_10X, LIHC_GSE140228_Smartseq2, and LIHC_GSE98638) were retrieved from TISCH database (http://tisch.comp-genomics.org/) (44). E2F1 expression levels in different major-lineage cell clusters of each dataset were investigated. Moreover, the hallmark of up-regulated genes in different cell clusters, as well as the enrichment of E2F targets were also explored via TISCH database. Furthermore, the correlations between E2F1 expression and the infiltrations of different types of immune cells in LIHC were analyzed by TIMER2.0 database.
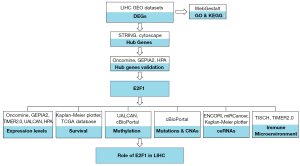
The brief study process was illustrated by flowchart (see Figure 1).
Statistical analysis
All statistical analysis were performed by SPSS Version 26. Multivariate Cox regression analysis was carried out to identify independent prognostic factors for LIHC. Simple linear regression was performed to calculate the correlation between promotor methylation and mRNA expression of E2F1, as well as relative linear copy-number values and mRNA expression of E2F1. Besides, E2F1 mRNA expression levels in different copy number groups were compared with One-way ANOVA. P value <0.05 is considered statistically significant, the confidence interval was set at 95%.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Overexpressed genes in LIHC
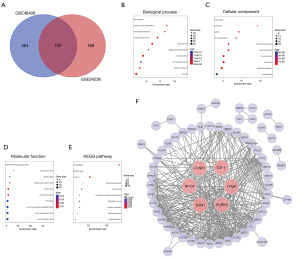
A total of 664 and 289 overexpressed DEGs were identified from GSE46408 and GSE54238 respectively. Among those, 100 overexpressed DEGs shared in these 2 GEO series were selected for further investigation (see Figure 2A).
Gene ontology and KEGG pathway enrichment
Analyzed by WebGestalt, the biological process annotation of the 100 overexpressed DEGs mainly enriched in DNA replication, mitotic cell cycle phase transition, cell cycle phase transition, mitotic cell cycle process, mitotic cell cycle, regulation of cell cycle process, cell cycle process, DNA metabolic process, regulation of cell cycle, and cell cycle (see Figure 2B). Cellular component mainly focused on mitotic spindle, spindle pole, spindle, chromosomal region, nuclear chromosome part, nuclear chromosome, chromosomal part, chromosome, microtubule cytoskeleton, and cytoskeletal part (see Figure 2C). Molecular function was mainly related to RNA-DNA hybrid ribonuclease activity, DNA helicase activity, helicase activity, catalytic activity, acting on DNA, kinase binding, protein kinase binding, ATP binding, purine ribonucleoside triphosphate binding, purine ribonucleotide binding, and purine nucleotide binding (see Figure 2D). KEGG pathway mainly concentrated on DNA replication, cell cycle, and oocyte meiosis (see Figure 2E).
PPI network construction and hub genes selection
Sixty-four largest connected nodes (genes) and 375 edges were constructed through STRING and illustrated by Cytoscape. The other 36 nodes (genes) belonging to separate interaction cluster were not included. Moreover, by using CytoHubba plugin, 6 hub genes named CCNB1, RFC4, E2F1, DSN1, TYMS, and AURKA were identified (see Figure 2F).
Hub genes validation
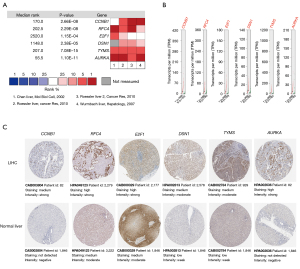
Four datasets containing the expression levels of numerous genes including 6 hub genes were selected and analyzed in Oncomine. Each hub gene was significantly overexpressed in LIHC in at least 2 out of 4 datasets and no hub gene was downregulated in these 4 datasets (see Figure 3A). Analysis by GEPIA2 also demonstrated that the mRNA expression level of each hub gene in LIHC was statistically higher than that in normal liver tissue (see Figure 3B). The immunohistochemical staining images showed that the protein expression of each hub gene exhibited higher level in LIHC samples than that in normal liver tissue (see Figure 3C).
E2F1 mRNA and protein expression in various cancers including LIHC
To deepen the understanding, this study selected one hub gene named E2F1 for further investigation. Firstly, the mRNA expression of E2F1 in various cancers and paired normal tissues were investigated. Analysis via Oncomine revealed that E2F1 was overexpressed in most solid tumors, such as liver cancer, lung cancer, breast cancer, colorectal cancer, kidney cancer, ovarian cancer, etc., except for brain and central nervous system cancer (see Figure 4A). Exploration by GEPIA2 found that E2F1 was significantly overexpressed in 23 out 31 kinds of solid tumors including LIHC, and no type of cancer exhibited downregulation of E2F1 (see Figure 4B). These findings were verified by investigating TIMER2.0, which showed that E2F1 was statistically overexpressed in 20 kinds of cancers containing LIHC (see Figure 4C).
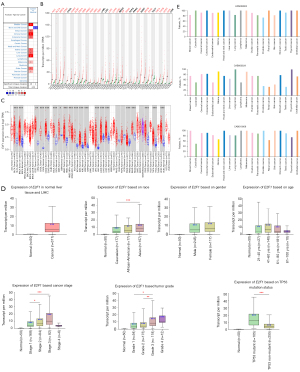
Furthermore, through analyzing 371 LIHC tissues and 50 normal tissues, UALCAN showed that E2F1 mRNA was overexpressed in LIHC regardless of race, gender, age, stage (except for stage 4), grade, and TP53 mutation status (see Figure 4D). Besides, stage 2 and 3, grade 3, as well as TP53 mutant subgroups exhibited higher E2F1 expression level compared to stage 1, grade 1 and 2, TP53 non-mutant subgroups respectively (see Figure 4D). In addition, HPA database showed that most kinds of cancers including liver cancer displayed moderate to strong immunoreactivity to E2F1 irrespective of different antibodies (see Figure 4E). To sum up, those data indicated that E2F1 mRNA and protein were overexpressed in most solid tumors including LIHC.
The relationship between E2F1 expression and survival in LIHC
Kaplan-Meier plotter was utilized to investigate survival data in accordance with E2F1 expression level. Results revealed that OS, DSS, PFS, and RFS were significantly in favor of E2F1 low expression cohort, and the hazard ratios (HR) were 1.75 (1.23–2.49, P=0.0016), 1.91 (1.21–3.01, P=0.0046), 1.60 (1.19–2.15, P=0.0018), and 1.68 (1.21–2.35, P=0.002) respectively (see Figure 5A-5D).
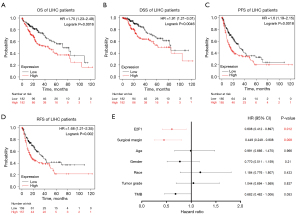
Moreover, subgroup analysis according to different clinicopathological characteristics indicated that the OS was significantly negatively correlated with E2F1 expression level in the following subgroups: male, Asian, Caucasian, stage 1, stages 1 and 2, stages 2 and 3, grade 2, no hepatitis virus infection, no alcohol consumption (see Figure S1A-S1I). In general, the results indicated that high levels of E2F1 were associated with poor prognosis in LIHC.
In addition, 366 LIHC patients were retrieved from TCGA database. Nineteen cases were excluded due to lack of complete data. Therefore, 347 cases were subject to analysis. Multivariate Cox regression analysis indicated that E2F1 expression level and surgical margin are independent factors influencing OS for LIHC. Specifically, low E2F1 expression level (HR =0.608, 95% CI: 0.412–0.897, P=0.012) and negative surgical margin (HR =0.449, 95% CI: 0.249–0.808, P=0.008) indicated better overall survival. Other factors such as age, gender, race, tumor grade, TMB (tumor mutation burden) were not related to prognosis (see Figure 5E).
E2F1 promotor methylation level in LIHC
To explore factors contributing to E2F1 overexpression in LIHC, promotor methylation was firstly investigated from TCGA database via UALCAN web. Results showed that there was no significant difference of E2F1 promotor methylation level between LIHC and normal liver tissue, and were generally very low (see Figure 6A). More specifically, E2F1 promotor methylation level exhibited no statistical difference regardless of race, gender, age, stage, grade, nodal metastasis status, and TP53 mutation status. Nevertheless, the simple linear regression showed that E2F1 promotor methylation level could negatively affect E2F1 mRNA expression level in LIHC with the intercept of 1.362 and the slope of −2.069 (ANOVA P value <0.001, Coefficients P value <0.001) (see Figure 6B).
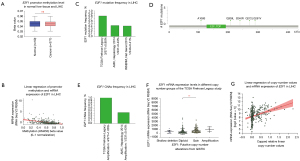
Mutations and CNAs of E2F1 in LIHC
Data indicated that E2F1 mutation frequency is low in LIHC. Only 0.41% to 0.80% of the LIHC samples contained mutations in the E2F1 gene (see Figure 6C). Five missense mutations were identified, including A108S, Q208L, S240R, Q272L, and G291V. All these 5 mutations were not located in the E2F/DP family winged-helix DNA-binding domain and the significance was unknown. No other mutations such as truncating mutations, in-frame mutations, splice mutations, or fusion mutations were found (see Figure 6D).
The frequency of CNAs ranged from 0.43% to 1.06% (see Figure 6E). Through analyzing 360 samples from the TCGA Firehose Legacy Study, it is indicated that E2F1 mRNA expression level was significantly higher in gain group than that in diploid group (P value =0.005) (see Figure 6F). In spite of no statistical significance due to small sample size, amplification group also showed higher E2F1 mRNA expression level compared to other groups (see Figure 6F). Moreover, the simple linear regression showed that relative linear copy-number values could positively affect E2F1 mRNA expression level in LIHC with the intercept of 8.239 and the slope of 1.567 (ANOVA P value <0.001, Coefficients P value <0.001) (see Figure 6G).
Possible ceRNAs of E2F1
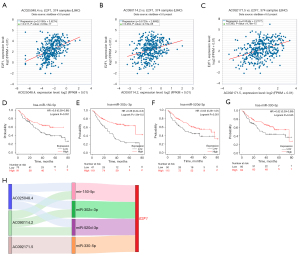
Since lncRNAs could negatively affect miRNAs which then negatively affect E2F1 levels, those lncRNAs and miRNAs positively and negatively correlated with E2F1 expression level respectively could be considered as ceRNAs. ENCORI showed that 8 lncRNAs are considered as possible ce-lncRNAs. Three out of 8 lncRNAs showed significantly positive correlation with E2F1 expression, named AC025048.4, AC090114.2, and AC092171.5 (see Figure 7A-7C). Thus, these 3 lncRNAs are supposed to be ce-lncRNAs. Twenty-six miRNAs that could be regulated by the above 3 ce-lncRNAs were found. Thirteen out of 26 miRNAs exhibited lower expression in LIHC than in normal liver tissue. Moreover, 4 out of 13 miRNAs displayed favorable OS in LIHC patients and were included into ceRNAs network, namely miR-150-5p, miR-302c-3p, miR-520d-3p, and miR-330-5p (see Figure 7D-7G). Thus, the ceRNAs network was established (see Figure 7H).
The association between E2F1 and tumor immune microenvironment
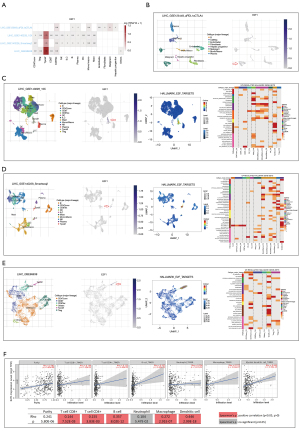
Through exploring single cell sequencing data via TISCH, 1 dataset (LIHC_GSE125449_aPDL1aCTLA4) including malignant cell cluster showed that E2F1 was expressed in malignant cell cluster (see Figure 8A,8B). The other 3 datasets including various immune cell clusters showed that E2F1 was mainly expressed in proliferating T cells (see Figure 8A,8C-8E). Gene set enrichment analysis also indicated that the hallmark of upregulated genes of proliferating T cells mainly focused on E2F targets, G2M checkpoints, mitotic spindle, and MYC targets (see Figure 8C-8E). Notably, E2F targets were almost exclusively enriched in proliferating T cells (see Figure 8C-8E). Furthermore, E2F1 was positively correlated with specific types of immune cells, including CD8(+) T cells, CD4(+) T cells, B cells, macrophages, dendritic cells (see Figure 8F). Therefore, these findings suggest that E2F1 could affect immune functions and thereby might influence the tumorigenesis and the development of LIHC.
Discussion
LIHC is one of the most lethal cancers. Most of LIHC patients are diagnosed at advanced stage and have poor prognosis with roughly equivalent incidence rate and mortality rate (2,45). It is indicated that some inner molecular mechanisms may play important roles leading to relatively high malignancy of LICH. Thus, exploring molecular mechanisms is of potential significance for a deeper understanding into LIHC. Nevertheless, although studies have unveiled substantial amounts of knowledge regarding mutations, epigenetic drivers, aberrant gene-expression profiles, disrupted signaling pathways and chromosomal aberrations (46), the efficacy of treatments regarding those mechanisms remains limited, and it desperately awaits a better understanding of the cancer’s molecular biology (47). Hereby, this study adopted bioinformatics to merge a great amount of data to explore the possible roles of E2F1 in LIHC.
The reasons for choosing E2F1 as the research objective are as follows: E2F1 as one of hub genes in LIHC was demonstrated by this study and was verified by several other studies (48,49). Moreover, the roles of E2F1 in tumorigenesis were investigated in other cancers. For instance, E2F1 was overexpressed in breast cancer and facilitated stemness and tumorigenesis through binding with the promoter region of Nanog gene and promoting its transcription (50). E2F1 mRNA expression level is related to pathological parameters and is an independently unfavorable factor for OS in clear cell renal cell carcinoma (51). Although E2F1 is one of the most researched E2Fs family members (52), the exact role of E2F1 in LIHC remains controversial and unsettled. It is reported that E2F1 is upregulated in LICH and promotes the development of LIHC via targeting MYBL2 or through being regulated by SIRT5 (15,53). E2F1 might also function as a critical anti-apoptotic factor by counteracting c-myc-initiated apoptosis via concomitant induction of PIK3CA/Akt/mTOR and c-Myb/COX-2 pathways (18). On the other hand, E2F1 could inhibit hepatitis B virus (HBV) associated LIHC by suppressing HBV life cycle through interfering with the control of HBx on p53 promoter as well as directly binding and activating p53 promoter (54). Also, E2F1 expression level proved to be positively related to tumor apoptotic index in LIHC (55). Since the proliferative and suppressive functions of E2F1 in LIHC may coexist, it is worth exploring the relationship between E2F1 and LIHC from translationally and clinically relevant viewpoints.
Hence, this study assessed the clinical significance of E2F1 in LIHC using a variety of databases to consolidate the results. It is confirmed that E2F1 mRNA and protein were overexpressed in LIHC regardless of clinicopathological characteristics. Moreover, those harboring more malignant features, such as advanced stage and high grade, exhibited higher E2F1 expression level, potentially indicate that E2F1 may serve as a vital molecule in LIHC development. Furthermore, this study revealed that E2F1 overexpression is associated with poor OS and PFS, denoting that E2F1 may be employed as an unfavorable prognostic factor for LIHC. These findings are, to some extent, in accordance with the standpoint of Farra et al. (56), proposing that the proliferative and apoptotic functions of E2F1 in LIHC may coexist but the proliferative effect seems to be more pronounced than the apoptotic one.
Possible mechanisms contributing to the upregulation of E2F1 were probed. Previous studies reported that E2F1 showed methylation differences between LIHC and healthy liver tissue, as well as between LIHC and cirrhosis tissue, suggesting E2F1 promotor methylation plays an important role in hepatocarcinogenesis (57,58). Gao et al. (59,60) suggested that E2F1 could be epigenetically regulated by PcG via EZH2-mediated H3K27me3 in LIHC. However, this study indicated promotor methylation may not be considered as a vital factor that could influence E2F1 expression.
Gene amplification is also closely linked to the pathogenesis and anti-tumor drug sensitivity in various cancers (61). Chromosome 1q21-23 amplification has been identified as the most frequent chromosomal alteration associated with LIHC (62). Besides, CCN2/MAPK/Id-1 loop feedback amplification is involved in oxaliplatin resistance, and CCN2 or MAPK signaling inhibitor could ameliorate oxaliplatin resistance in LIHC (63). This study revealed amplification is one of the contributing factors to the overexpression of E2F1. Kent et al. have shown that copy number gains in E2F1 or E2F3b resulted in dosage-dependent spontaneous LIHC in mice without the involvement of additional organs (64), suggesting that E2F1 is a vital molecule for LIHC.
Dysregulation of ceRNAs network has been implicated in the pathogenesis of cancers like LIHC (65). An LIHC-specific lncRNA–miRNA–mRNA ceRNAs network is being expanded in recent years and is deemed as a potential prognostic marker (66). It is also found that ceRNAs could be harnessed to support LIHC treatment and improve drug sensitivity (65,67). Nonetheless, the ceRNAs network of E2F1 in LIHC has not been well established. Here, a ceRNAs network of E2F1 including 3 lncRNAs and 4 miRNAs was mapped out. The 3 lncRNAs named AC025048.4, AC092171.5, and AC090114.2 were reported to be associated with poor prognosis in other cancers such as lung adenocarcinoma, pancreatic cancer, and cholangiocarcinoma, and the first two are related to immunoregulatory pathways (68-71). The carcinostatic role of the 4 miRNAs including miR-150-5p, miR-302c-3p, miR-520d-3p, and miR-330-5p in LIHC were verified by wet-lab experiment (72-75). Besides, miR-150-5p is supposed to be one of the segments regarding sorafenib resistance in LIHC (76).
Liver is a special immune-tolerant organ that can effectively escape the immune response (77), while the LIHC neoplasm contains a large number of immune cells, forming a complex immune microenvironment (78,79). The recent years have witnessed the emerging and unprecedent progression of immunotherapy, however, low response rate and consequent drug-resistance have largely limited the efficacy of immunotherapy (80). Exploring immune microenvironment to improve the efficacy of immunotherapy is needed. By analyzing single cell sequencing data, this study revealed that E2F1 was not only expressed in malignant cell cluster but also expressed in specific immune cell clusters. Yim et al. revealed that LIHC from transgenic mice expressing Myc/E2f1 or E2f1 exhibited poor prognosis. Moreover, 88% of LIHC from E2f1 model were classified into high immune activity subgroup with high Pd-1 expression, which can predict the response to Pembrolizumab (81). Mechanically, one study indicated that CASC11 and E2F1 could impact the activation of the NF-κB signaling and PI3K/Akt/mTOR pathway and further regulate the expression PD-L1 (82). Together with our findings, those evidence suggests that E2F1 could affect immune microenvironment and thereby might be employed as a potential therapeutic target for immunotherapy.
E2F1/E2F4 inhibitor HLM006474 has been preclinically applied to treat several kinds of cancers. Rouaud et al. (83) demonstrated that HLM006474 had an anti-proliferative effect on melanoma cells. Some relevant E2F1 target genes, such as BIRC5, CDC6, or BRCA1 were inhibited by HLM006474. Moreover, HLM006474 could induce cell cycle arrest in the G2/M phase, apoptosis via caspase-3 and p53, and senescence. Meanwhile, it proved that HLM006474 could reduce the viability of lung cancer cell lines (84) and potentially alter the progression of myelodysplastic syndrome into acute myeloid leukemia (85). Meanwhile, Sheldon (86) indicated that in breast cancer, arsenic trioxid could inhibit the dissociation of E2F1 from the tumor suppressor, retinoblastoma protein (pRB) due to changes in pRB phosphorylation which leads to decreased E2F1 transcriptional activity. There are also a few studies about E2F1 inhibitors for hepatocellular carcinoma. The small molecule BTYNB could interfere with E2F-driven gene expression and tumor growth in experimental mouse tumor models (87). Liu et al. (88) reported that hepatocellular carcinoma tissues with embryonic stem cell-like signature were sensitive to HLM006474. Although preliminary data strengthen the hypothesis that E2F inhibitors could be useful as a new therapeutic approach for hepatocellular carcinoma, there is still a long way to go prior to clinical application.
Notably, one limitation of this study is that only bioinformatic methods were adopted to integrate and analyze numerous data. Further wet-lab verification is needed to make the findings more solid.
Conclusions
Bioinformatic analysis indicated that, as a hub gene, E2F1 was overexpressed in LIHC and related to poor prognosis. Amplification and ceRNAs regulation may contribute to the overexpression of E2F1. E2F1 could affect immune microenvironment. Together, this study indicated that E2F1 is one of the key genes and could be used as a potential prognostic biomarker and therapeutic target for LIHC.
Acknowledgments
We would like to thank Peng Jiang for English language proofreading.
Funding: This work was supported by the Natural Science Foundation of Guangdong Province (No. 2018A030313530 to ZT); the Chinese Scholarship Council (No. 201908440010 to ZT); the Shenzhen Science and Technology Innovation Commission Project (No. JCYJ20210324110408022 to ZT; Nos. GJHZ20180420180754917, ZDSYS20190902092855097, KCXFZ20200201101050887 and GJHZ20200731095207023 to YL); the Beijing Bethune Charitable Foundation (No. flzh202120 to ZT); and the Shenzhen Sanming Project (No. SZSM201612041 to YL).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-218/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-218/coif). ZT reports grants from the Natural Science Foundation of Guangdong Province (No. 2018A030313530); the Chinese Scholarship Council (No. 201908440010); the Shenzhen Science and Technology Innovation Commission Project (No. JCYJ20210324110408022); and the Beijing Bethune Charitable Foundation (No. flzh202120). YL reports grants from the Shenzhen Science and Technology Innovation Commission Project (Nos. GJHZ20180420180754917, ZDSYS20190902092855097, KCXFZ20200201101050887 and GJHZ20200731095207023); and the Shenzhen Sanming Project (No. SZSM201612041). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021;73:4-13. [Crossref] [PubMed]
- Llovet JM, Montal R, Sia D, et al. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15:599-616. [Crossref] [PubMed]
- Ding J, Wen Z. Survival improvement and prognosis for hepatocellular carcinoma: analysis of the SEER database. BMC Cancer 2021;21:1157. [Crossref] [PubMed]
- Anwanwan D, Singh SK, Singh S, et al. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer 2020;1873:188314. [Crossref] [PubMed]
- Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6. [Crossref] [PubMed]
- Morgunova E, Yin Y, Jolma A, et al. Structural insights into the DNA-binding specificity of E2F family transcription factors. Nat Commun 2015;6:10050. [Crossref] [PubMed]
- Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer 2019;19:326-38. [Crossref] [PubMed]
- Wang H, Wang X, Xu L, et al. Integrated analysis of the E2F transcription factors across cancer types. Oncol Rep 2020;43:1133-46. [Crossref] [PubMed]
- Yang M, Wu S, Jia J, et al. JAZ mediates G1 cell cycle arrest by interacting with and inhibiting E2F1. Cell Cycle 2011;10:2390-9. [Crossref] [PubMed]
- Jiang X, Nevins JR, Shats I, et al. E2F1-Mediated Induction of NFYB Attenuates Apoptosis via Joint Regulation of a Pro-Survival Transcriptional Program. PLoS One 2015;10:e0127951. [Crossref] [PubMed]
- Li S, Yang X, Li W, et al. Comprehensive Analysis of E2F Family Members in Human Gastric Cancer. Front Oncol 2021;11:625257. [Crossref] [PubMed]
- Huang YL, Ning G, Chen LB, et al. Promising diagnostic and prognostic value of E2Fs in human hepatocellular carcinoma. Cancer Manag Res 2019;11:1725-40. [Crossref] [PubMed]
- Qiao L, Zhang Q, Sun Z, et al. The E2F1/USP11 positive feedback loop promotes hepatocellular carcinoma metastasis and inhibits autophagy by activating ERK/mTOR pathway. Cancer Lett 2021;514:63-78. [Crossref] [PubMed]
- Nakajima T, Yasui K, Zen K, et al. Activation of B-Myb by E2F1 in hepatocellular carcinoma. Hepatol Res 2008;38:886-95. [Crossref] [PubMed]
- Chen Q, Wang L, Jiang M, et al. E2F1 interactive with BRCA1 pathway induces HCC two different small molecule metabolism or cell cycle regulation via mitochondrion or CD4+T to cytosol. J Cell Physiol 2018;233:1213-21. [Crossref] [PubMed]
- Chen YL, Uen YH, Li CF, et al. The E2F transcription factor 1 transactives stathmin 1 in hepatocellular carcinoma. Ann Surg Oncol 2013;20:4041-54. [Crossref] [PubMed]
- Ladu S, Calvisi DF, Conner EA, et al. E2F1 inhibits c-Myc-driven apoptosis via PIK3CA/Akt/mTOR and COX-2 in a mouse model of human liver cancer. Gastroenterology 2008;135:1322-32. [Crossref] [PubMed]
- Sun HX, Xu Y, Yang XR, et al. Hypoxia inducible factor 2 alpha inhibits hepatocellular carcinoma growth through the transcription factor dimerization partner 3/ E2F transcription factor 1-dependent apoptotic pathway. Hepatology 2013;57:1088-97. [Crossref] [PubMed]
- Forman MR, Greene SM, Avis NE, et al. Bioinformatics: Tools to accelerate population science and disease control research. Am J Prev Med 2010;38:646-51. [Crossref] [PubMed]
- Chen YL, Wang TH, Hsu HC, et al. Overexpression of CTHRC1 in hepatocellular carcinoma promotes tumor invasion and predicts poor prognosis. PLoS One 2013;8:e70324. [Crossref] [PubMed]
- Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016;63:499-511. [Crossref] [PubMed]
- Yuan S, Wang J, Yang Y, et al. The Prediction of Clinical Outcome in Hepatocellular Carcinoma Based on a Six-Gene Metastasis Signature. Clin Cancer Res 2017;23:289-97. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Liao Y, Wang J, Jaehnig EJ, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 2019;47:W199-205. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Thul PJ, Åkesson L, Wiking M, et al. A subcellular map of the human proteome. Science 2017;356:eaal3321. [Crossref] [PubMed]
- Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci 2018;5:181006. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Ahn SM, Jang SJ, Shim JH, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014;60:1972-82. [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Ala U. Competing Endogenous RNAs, Non-Coding RNAs and Diseases: An Intertwined Story. Cells 2020;9:1574. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Xie B, Ding Q, Han H, et al. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics 2013;29:638-44. [Crossref] [PubMed]
- Sun D, Wang J, Han Y, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res 2021;49:D1420-30. [Crossref] [PubMed]
- Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol 2020;72:250-61. [Crossref] [PubMed]
- Raja A, Haq F. Molecular classification of hepatocellular carcinoma: prognostic importance and clinical applications. J Cancer Res Clin Oncol 2022;148:15-29. [Crossref] [PubMed]
- Juaid N, Amin A, Abdalla A, et al. Anti-Hepatocellular Carcinoma Biomolecules: Molecular Targets Insights. Int J Mol Sci 2021;22:10774. [Crossref] [PubMed]
- Liu Z, Pu Y, Bao Y, et al. Investigation of Potential Molecular Biomarkers for Diagnosis and Prognosis of AFP-Negative HCC. Int J Gen Med 2021;14:4369-80. [Crossref] [PubMed]
- Liu J, Li W, Zhang J, et al. Identification of key genes and long non-coding RNA associated ceRNA networks in hepatocellular carcinoma. PeerJ 2019;7:e8021. [Crossref] [PubMed]
- Lu G, Li Y, Ma Y, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res 2018;37:289. [Crossref] [PubMed]
- Ma X, Gao Y, Fan Y, et al. Overexpression of E2F1 promotes tumor malignancy and correlates with TNM stages in clear cell renal cell carcinoma. PLoS One 2013;8:e73436. [Crossref] [PubMed]
- Zhang C, Cui Y, Wang G, et al. Comprehensive Analysis of the Expression and Prognosis for E2Fs in Human Clear Cell Renal Cell Carcinoma. J Healthc Eng 2021;2021:5790416. [Crossref] [PubMed]
- Chang L, Xi L, Liu Y, et al. SIRT5 promotes cell proliferation and invasion in hepatocellular carcinoma by targeting E2F1. Mol Med Rep 2018;17:342-9. [PubMed]
- Choi M, Lee H, Rho HM. E2F1 activates the human p53 promoter and overcomes the repressive effect of hepatitis B viral X protein (Hbx) on the p53 promoter. IUBMB Life 2002;53:309-17. [Crossref] [PubMed]
- Palaiologou M, Koskinas J, Karanikolas M, et al. E2F-1 is overexpressed and pro-apoptotic in human hepatocellular carcinoma. Virchows Arch 2012;460:439-46. [Crossref] [PubMed]
- Farra R, Grassi G, Tonon F, et al. The Role of the Transcription Factor E2F1 in Hepatocellular Carcinoma. Curr Drug Deliv 2017;14:272-81. [PubMed]
- Jueliger S, Lyons J, Cannito S, et al. Efficacy and epigenetic interactions of novel DNA hypomethylating agent guadecitabine (SGI-110) in preclinical models of hepatocellular carcinoma. Epigenetics 2016;11:709-20. [Crossref] [PubMed]
- Fernández-Barrena MG, Arechederra M, Colyn L, et al. Epigenetics in hepatocellular carcinoma development and therapy: The tip of the iceberg. JHEP Rep 2020;2:100167. [Crossref] [PubMed]
- Gao SB, Xu B, Ding LH, et al. The functional and mechanistic relatedness of EZH2 and menin in hepatocellular carcinoma. J Hepatol 2014;61:832-9. [Crossref] [PubMed]
- Gao SB, Zheng QF, Xu B, et al. EZH2 represses target genes through H3K27-dependent and H3K27-independent mechanisms in hepatocellular carcinoma. Mol Cancer Res 2014;12:1388-97. [Crossref] [PubMed]
- Shimizu N. Gene Amplification and the Extrachromosomal Circular DNA. Genes (Basel) 2021;12:1533. [Crossref] [PubMed]
- Steinemann D, Skawran B, Becker T, et al. Assessment of differentiation and progression of hepatic tumors using array-based comparative genomic hybridization. Clin Gastroenterol Hepatol 2006;4:1283-91. [Crossref] [PubMed]
- Liao X, Bu Y, Jiang S, et al. CCN2-MAPK-Id-1 loop feedback amplification is involved in maintaining stemness in oxaliplatin-resistant hepatocellular carcinoma. Hepatol Int 2019;13:440-53. [Crossref] [PubMed]
- Kent LN, Bae S, Tsai SY, et al. Dosage-dependent copy number gains in E2f1 and E2f3 drive hepatocellular carcinoma. J Clin Invest 2017;127:830-42. [Crossref] [PubMed]
- Afify AY, Ibrahim SA, Aldamsisi MH, et al. Competing Endogenous RNAs in Hepatocellular Carcinoma-The Pinnacle of Rivalry. Semin Liver Dis 2019;39:463-75. [Crossref] [PubMed]
- Zhang J, Fan D, Jian Z, et al. Cancer Specific Long Noncoding RNAs Show Differential Expression Patterns and Competing Endogenous RNA Potential in Hepatocellular Carcinoma. PLoS One 2015;10:e0141042. [Crossref] [PubMed]
- Tang S, Tan G, Jiang X, et al. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget 2016;7:73257-69. [Crossref] [PubMed]
- Zheng Z, Zhang Q, Wu W, et al. Identification and Validation of a Ferroptosis-Related Long Non-coding RNA Signature for Predicting the Outcome of Lung Adenocarcinoma. Front Genet 2021;12:690509. [Crossref] [PubMed]
- Guo Y, Qu Z, Li D, et al. Identification of a prognostic ferroptosis-related lncRNA signature in the tumor microenvironment of lung adenocarcinoma. Cell Death Discov 2021;7:190. [Crossref] [PubMed]
- Wei C, Liang Q, Li X, et al. Bioinformatics profiling utilized a nine immune-related long noncoding RNA signature as a prognostic target for pancreatic cancer. J Cell Biochem 2019;120:14916-27. [Crossref] [PubMed]
- Xie X, Wang Y, Zhang S, et al. A novel five-lncRNA signature panel improves high-risk survival prediction in patients with cholangiocarcinoma. Aging (Albany NY) 2021;13:2959-81. [Crossref] [PubMed]
- Cui X, Jiang X, Wei C, et al. Astragaloside IV suppresses development of hepatocellular carcinoma by regulating miR-150-5p/β-catenin axis. Environ Toxicol Pharmacol 2020;78:103397. [Crossref] [PubMed]
- Yang L, Guo Y, Liu X, et al. The tumor suppressive miR-302c-3p inhibits migration and invasion of hepatocellular carcinoma cells by targeting TRAF4. J Cancer 2018;9:2693-701. [Crossref] [PubMed]
- Xiang Y, Huang Y, Sun H, et al. Deregulation of miR-520d-3p promotes hepatocellular carcinoma development via lncRNA MIAT regulation and EPHA2 signaling activation. Biomed Pharmacother 2019;109:1630-9. [Crossref] [PubMed]
- Gong D, Feng PC, Ke XF, et al. Silencing Long Non-coding RNA LINC01224 Inhibits Hepatocellular Carcinoma Progression via MicroRNA-330-5p-Induced Inhibition of CHEK1. Mol Ther Nucleic Acids 2020;19:482-97. [Crossref] [PubMed]
- Sui C, Dong Z, Yang C, et al. LncRNA FOXD2-AS1 as a competitive endogenous RNA against miR-150-5p reverses resistance to sorafenib in hepatocellular carcinoma. J Cell Mol Med 2019;23:6024-33. [Crossref] [PubMed]
- Rizvi S, Wang J, El-Khoueiry AB. Liver Cancer Immunity. Hepatology 2021;73:86-103. [Crossref] [PubMed]
- Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018;68:1025-41. [Crossref] [PubMed]
- Nishida N, Kudo M. Immunological Microenvironment of Hepatocellular Carcinoma and Its Clinical Implication. Oncology 2017;92:40-9. [Crossref] [PubMed]
- Khan AA, Liu ZK, Xu X. Recent advances in immunotherapy for hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int 2021;20:511-20. [Crossref] [PubMed]
- Yim SY, Shim JJ, Shin JH, et al. Integrated Genomic Comparison of Mouse Models Reveals Their Clinical Resemblance to Human Liver Cancer. Mol Cancer Res 2018;16:1713-23. [Crossref] [PubMed]
- Song H, Liu Y, Li X, et al. Long noncoding RNA CASC11 promotes hepatocarcinogenesis and HCC progression through EIF4A3-mediated E2F1 activation. Clin Transl Med 2020;10:e220. [Crossref] [PubMed]
- Rouaud F, Hamouda-Tekaya N, Cerezo M, et al. E2F1 inhibition mediates cell death of metastatic melanoma. Cell Death Dis 2018;9:527. [Crossref] [PubMed]
- Kurtyka CA, Chen L, Cress WD. E2F inhibition synergizes with paclitaxel in lung cancer cell lines. PLoS One 2014;9:e96357. [Crossref] [PubMed]
- He Z, Zhang S, Ma D, et al. HO-1 promotes resistance to an EZH2 inhibitor through the pRB-E2F pathway: correlation with the progression of myelodysplastic syndrome into acute myeloid leukemia. J Transl Med 2019;17:366. [Crossref] [PubMed]
- Sheldon LA. Inhibition of E2F1 activity and cell cycle progression by arsenic via retinoblastoma protein. Cell Cycle 2017;16:2058-72. [Crossref] [PubMed]
- Müller S, Bley N, Busch B, et al. The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res 2020;48:8576-90. [Crossref] [PubMed]
- Liu M, Yan Q, Sun Y, et al. A hepatocyte differentiation model reveals two subtypes of liver cancer with different oncofetal properties and therapeutic targets. Proc Natl Acad Sci U S A 2020;117:6103-13. [Crossref] [PubMed]


