
Clinicopathological and prognostic significance of the long non-coding RNA HOTAIR high expression in head and neck squamous cell carcinoma: a systematic review and meta-analysis
Introduction
Head and neck tumors include carcinoma originating in any tissue or organ of the head and neck, with the exception of the brain, eyes, ears, thyroid, and esophagus (1). In addition, more than 90% of head and neck tumors are squamous cell carcinomas (2,3). Head and neck squamous cell carcinoma (HNSCC) is one of the most common malignant tumors in humans and includes a variety of tumors located in the mouth, nose, oropharynx, hypopharynx, and larynx (4,5). It is primarily caused by smoking, alcohol consumption, and human papillomavirus (HPV) infection (6,7). HNSCC has a high morbidity and mortality rate (8,9), and despite great advances in its prevention, diagnosis, and treatment, 5-year overall survival (OS) remains low due to lymphnode metastasis and local recurrence (10,11). In general, tumor clinical stage, lymphnode metastasis, and histological grades are directly related to HNSCC prognosis, but usually do not represent its true evolution. Therefore, to accurately predict HNSCC prognosis, develop therapeutic targets and conduct treatment evaluations, it is important to search for new valuable biomarkers (12).
Long non-coding RNAs (lncRNAs) are a group of RNAs over 200 nucleotides long, and are mostly transcribed by RNA polymerase II (13). LncRNA can play a role in cis and/or trans forms, most of which do not have any protein-coding capacity (14,15). LncRNA was previously defined as “junk” or “noise” (16,17). However, as time passed, lncRNAs have been found to have important biological functions in regulating transcription and gene expression, which can regulate disease occurrence and progression (18). Since they play an important role in all levels of gene expression, lncRNAs have attracted extensive attention in various physiological processes, such as differentiation and metabolism (19-21). Many scholars have studied the process of lncRNA involved in the occurrence and development of HNSCC, the results show that the up-regulation and down-regulation of lncRNA may be involved in the occurrence, development, invasion and metastasis of HNSCC (22,23). LncRNAs affect the clinicopathological features of tumors by participating in these processes and regulating epithelial-mesenchymal transition (EMT) (21,24). And the prognostic value of lncRNA expression in HNSCC remains to be studied.
In HNSCC, some lncRNAs, such as HOTAIR, have been posted to act a key function in tumor development by regulating cell proliferation (25), differentiation (26), and metastasis (27). HOX transcript antisense intergenic RNA (HOTAIR) interacts with the polycomb repressive complex 2 (PRC2) and then participates in the H3 lysine 27 (H3K27) methylation of many genes, which is associated with tumor physiological processes (28). HOTAIR is one of the most widely studied lncRNAs associated with human tumors. Compared with coding genes, lncRNA HOTAIR has clear tissue specificity (29), and exhibits unique characteristics in many types of cancer. HOTAIR is considered a significant biomarker of diagnoses and prognosis evaluations (30). Plus, they can be combined with clinicopathological characteristics to guide which therapeutic strategy to choose (31). Many studies have proven that HOTAIR is related to the occurrence, proliferation, invasion, and metastasis of different types of cancers, including breast cancer (32), lung cancer (33), gastric cancer (34), liver cancer (35), head and neck cancer (27), etc.
Although a number of studies are related to HOTAIR in HNSCC, no conclusive analysis with sufficient evidence has been formulated to summarize these existing studies (36-38). A previous meta-analysis showed that high HOTAIR expressions were associated with poor prognosis in HNSCC [pooled hazard ratio (HR) =1.90, 95% confidence interval (CI): 1.42–2.53, P<0.0001], but this prognostic analysis included only four studies with a total of 271 patients (39). In addition, only 2–3 of these studies evaluated the relationship between HOTAIR and tumor stages, histological grades, and lymph node metastasis. Their results showed that higher levels of HOTAIR expressions were associated with higher tumor stages and lymph node metastasis, but not with histologic grades. However, the number of studies is too small and the heterogeneity was not explained, so the reliability and accuracy of the results are insufficient.
Effect and regulation of HOTAIR on prognosis and clinical malignant phenotype in patients with HNSCC is still lacking convincing evidence-based medicine proof. Therefore, we reviewed the recent data on HOTAIR in relation to HNSCC. To explore the effects of lncRNAs HOTAIR on clinicopathological features and prognosis of HNSCC, meta-analysis, sensitivity analysis, and subgroup analysis were used in our study to draw more consistent and accurate conclusions. In addition, we added the disease-free survival (DFS) index in the prognostic analysis, and the tumor clinical stage was divided into T stages and TNM stages to analyse them separately. The addition and refinement of these indicators may help us better understand the role of HOTAIR in the occurrence, progression, and prognosis of HNSCC. Therefore, we did not define this study as an update of previous studies, but a new analysis with more research volume and more detailed study methods. We present the following article in accordance with the PRISMA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-652/rc) (40).
Methods
Search strategy
The research was implemented independently by two of the authors in the following online databases: PubMed, Web of Science, Embase, Cochrane Library and SCOPUS. The studies published before September 10, 2021 were screened. MESH terms and free text words were used to search for eligible studies on databases. The search strategy was: (HOTAIR OR “HOX transcript antisense RNA” OR “HOX transcript antisense intergenic RNA” OR “homeobox transcript antisense RNA”) AND (“head and neck” OR oropharyngeal OR oropharynx OR oral OR mouth OR gingival OR lip OR palatal OR tongue OR nasopharyngeal OR pharynx OR pharyngeal OR laryngeal OR larynx) AND (“squamous cell carcinoma” OR cancer OR carcinoma OR tumor OR neoplasm).
Inclusion and exclusion criteria
Inclusion criteria were: (I) only English language articles about cohort studies, but there were no restrictions on publication date, region, gender, age or follow-up period; (II) analysis of HOTAIR high expression associations with at least one of the following clinicopathological and prognostic variables in patients diagnosed with HNSCC: OS, DFS, T stage, lymphnode metastasis, TNM stage, histological grade; (III) in the initial literature screen, we found that the data of three pieces about the high expression of HOTAIR and the prognosis of HNSCC were derived from The Cancer Genome Atlas (TCGA) database. After comparison, only the more complete or more recent articles were selected to avoid including duplicated data; (IV) studies including quantitative analysis of tissue sections from surgery through quantitative PCR (qPCR), in situ hybridization (ISH), and fluorescence in situ hybridization (FISH); (V) study requirements for inclusion in survival analysis: all patients did not receive preoperative chemotherapy or radiotherapy, only radical surgery.
Exclusion criteria were: (I) studies with insufficient data; (II) reviews, case reports, small sample studies and conference abstracts; (III) neoplasms were derived from the salivary glands, thyroid and skin; (IV) in vitro models or animal experiments.
Data extraction
Data extraction was performed independently by two authors. The following parameters were extracted from each included study: name of the first author, year of publication, patient source, total number and high/low expressions of HOTAIR, tumor sites, detection methods of HOTAIR expressions, cut-off values, tumor clinicopathologic features (T stage, lymphnode metastasis, TNM stage, and histological grades), HRs and 95% CIs for OS and DFS, follow-up periods. If the articles only provide survival curves without directly providing HRs and 95% CIs, we obtained the HRs, 95% CIs and associated statistics from the survival curves or Cox regression analyses using Engauge Digitizer 4.1 software and the methods illustrated by Tierney et al. (41). During the data extraction process, any differences were resolved through discussion or based on the opinions of the third author.
Quality evaluation
Quality evaluation of the included studies was conducted independently by two researchers using the Newcastle-Ottawa Scale (NOS). Cohort study quality assessment of NOS contains eight domains, including representativeness of the exposed cohort, selection of the non-exposed cohort, ascertainment of exposure, demonstrations that outcome of interest was not present at the start of study, comparability of cohorts on the basis of the design or analysis, assessment of outcome, follow-ups long enough for outcomes to occur, adequacy of follow-up of cohorts. In each domain, 2–4 prompting items were assessed to conclude an overall evaluation.
Statistical analysis
Meta analysis was carried out using Review manager v5.3 software (Cochrane Collaboration, Oxford, United Kingdom) and Stata v11.0 software (Stata Corporation, College Station, Texas). Moreover, the relationship between HOTAIR expressions and clinicopathological characteristics was evaluated by referring to odds ratios (ORs) and 95% CIs. The features included tumor T-stage, lymph node metastasis, TNM stage, and histological grades. Meanwhile, HRs and 95% CIs were used to assess any association between HOTAIR expressions and HNSCC prognoses (OS and DFS). For studies that did not report accurate HRs and 95% CIs, Engauge Digitizer V4.1 software was used to extract results from Kaplan-Meier curves.
The presence of heterogeneity among studies was evaluated through Q-testing and P values. If there no evidence of significant heterogeneity (P>0.10 and/or I2<50%) was observed, a fixed-effects model was adopted. Otherwise, a random-effects model was used to calculate pooled HRs or ORs.
Galbraith diagrams were constructed to identify heterogeneity. In addition, sensitivity analyses were performed to test the reliability of the meta-analysis results. To further identify sources of heterogeneity and confirm the stability of the results, subgroup analyses were performed based on three aspects: patient source (China or India), cut-offs (median, mean or other), and detection methods (PCR or ISH). Finally, funnel plots were constructed, and publication bias was measured by Begg’s test. Results with P<0.05 were considered statistically significant.
Results
Study selection and features
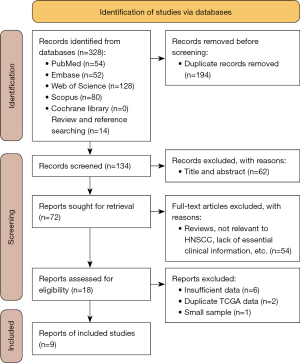
As shown in Figure 1, the literature search yielded a total of 328 studies from the databases (PubMed, Embase, Scopus, Web of Science), and the preliminary list screening (reviews and references). A total of 134 studies remained when duplicates were excluded. Then, 62 records were excluded after reading the titles and abstracts, and the remaining 72 were screened for full-text reading. After reading the full text, 18 studies were identified as eligible articles. According to the inclusion and exclusion criteria, 9 pieces of literature were eventually included in this study (26,36-38,42-46).
All of the included studies were published between 2012 and 2021 with sample sizes between 44 and 425 patients and a total of 1,043 patients. Seven studies with a total of 546 patients provided an association between the clinicopathological characteristics of HNSCC patients and HOTAIR expressions (26,36-38,43-45), and 6 studies with a total of 856 patients provided the correlation between HOTAIR expressions and HNSCC prognoses (26,36,38,42,45,46). Among the 9 studies, 7 studies were conducted in China, 1 in India (37), and 1 study involving survival outcomes was derived from TCGA database (46). Table 1 and Table S1 shows the main characteristics of these included studies.
Table 1
| Study | Year | Source | Total | High/low | Sample | Tumor site | Detection method | Follow-up (months) | Cut-off | Outcome | HR | 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cheng-Zhi Xu (36) | 2016 | China | 73 | 31/42 | Tissue | Head and neck | PCR | 250 | NA | OS | 1.43 | 0.66–3.10 |
| Dandan Li (42) | 2013 | China | 72 | 33/39 | Tissue | Laryngeal | PCR | 60 | NA | OS | 2.856 | 1.154–7.071 |
| Detao Tao (43) | 2020 | China | 44 | 22/22 | Tissue | Oral | PCR | – | Median | – | – | – |
| Fenglian Yang (44) | 2021 | China | 83 | 46/37 | Tissue | Nasopharynx | PCR | – | Mean | – | – | – |
| Ganesan Arunkumar (37) | 2017 | India | 60 | 12/43 | Tissue | Oral | PCR | – | Mean | – | – | – |
| Jie Wu (45) | 2015 | China | 50 | 25/25 | Tissue | Oral | PCR | 8–60 (median: 37) | Median | OS | 1.47 | 0.43–4.98 |
| Madeleine Sassenberg (46) | 2019 | TCGA | 425 | 58/367 | NA | Head and neck | NA | Median: 23 | NA | OS | 2.38 | 1.60–3.55 |
| Yan Nie (38) | 2013 | China | 160 | 91/69 | Tissue | Nasopharynx | ISH | Median: 69 | High (SI ≥6); low (SI <6) | DFS; OS | DFS: 1.7; OS: 1.56 | DFS: 1.04–2.78; OS: 0.80–3.02 |
| Yansheng Wu (26) | 2015 | China | 76 | 38/38 | Tissue | Oral | PCR | 100 | Median | DFS; OS | DFS: 1.52; OS: 1.91 | DFS: 0.73–3.15; OS: 0.70–5.22 |
CI, confidence interval; DFS, disease-free survival; HR, hazard ratio; ISH, in situ hybridization; SI, staining index; NA, not applicable; OS, overall survival; TCGA, The Cancer Genome Atlas database.
All the included literature was observational retrospective studies. The bias risks of the included studies were evaluated according to the NOS scale. Among the 9 included pieces of literature, 4 received 8 stars, 3 received 7 stars, 1 received 6 stars (the study involved only clinicopathological features and no follow-ups), and 1 received 5 stars (the data was derived from the TCGA database, and some of the information was not clear), all of which exceeded 5 stars. The specific scores of the included studies are shown in the Table S2. Most of the included studies had NOS scores of 7 to 8, indicating that these studies were of high quality.
Associations between lncRNA HOTAIR and clinicopathological characteristics
T stages
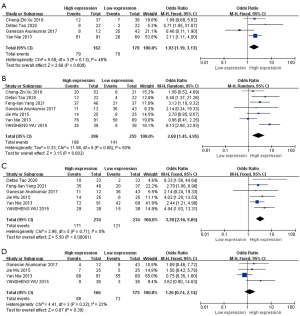
We compared the expressions of HOTAIR at different T stages in patients with HNSCC from 4 studies with 332 patients (36-38,43). Among the patients with T3–T4 stages, 79 had high HOTAIR expressions, and 70 cases exhibited low expressions for HOTAIR. A significant association was found between HOTAIR high expression and advanced T stages in HNSCC (OR =1.93, 95% CI: 1.19–3.13, P=0.008) (Figure 2A). Heterogeneity was moderate (I2=46%, P=0.13). As P=0.13 for the Q test, it was suggested that the heterogeneity among the selected studies was not statistically significant, and fixed effects could be selected for meta-analysis.
According to the Begg’s bias test based on the funnel plot (Figure S1A), P=1.000 was obtained, indicating that there were no publication biases in the four studies.
Lymphnode metastasis
Next, HOTAIR expression was compared among patients with HNSCC with different Lymphnode metastasis statuses from 7 studies that included in 541 patients (26,36-38,43-45). Among the patients with lymphnode metastasis (N1–N3), 196 patients exhibited high expressions for HOTAIR and 141 patients exhibited low expressions.
The results revealed that high expressions for HOTAIR had a significant association with lymphnode metastasis (OR =2.68, 95% CI: 1.45–4.95, P=0.002) (Figure 2B). Significant heterogeneity was found among the studies (I2=50%, P=0.06); thus, the pooled analysis was estimated using a random-effects model.
Through the Begg’s bias test, P=0.764, indicating that there were no publication biases in the 6 pieces of literature (Figure S1B).
TNM stages
In total, 468 patients across 6 studies were assessed to determine the relationships between TNM stages and HOTAIR expressions (26,37,38,43-45). Among the patients with high TNM stages (III–IV), 171 exhibited high HOTAIR expressions, and 121 exhibited low HOTAIR expressions.
The results revealed that HOTAIR high expression was significantly related to advanced TNM stages (OR =3.30, 95% CI: 2.16–5.05, P<0.00001) (Figure 2C). No heterogeneity was discovered between studies (I2=0%, P=0.71); therefore, the fixed-effect model was applied for pooled analyses.
As shown in the Funnel plot (Figure S1C), the Begg’s test was used to conduct publication bias. The Begg’s test analysis indicated there was no publication bias in these 6 studies due to the value of P=0.260.
Histological grades
The relationship between tumor histological grades and HOTAIR expressions was compared across 341 patients from 4 studies (26,37,38,45). Among the patients with poor histological grades, 88 had high HOTAIR expressions, and 72 had low HOTAIR expressions. However, the difference was not statistically significant (OR =1.26, 95% CI: 0.74–2.14, P=0.39) (Figure 2D). No apparent heterogeneity was found in the studies (I2=32%, P=0.22); therefore, the fixed-effect model was applied for pooled analysis.
As shown in the Figure funnel plot (Figure S1D), the Begg’s test indicated there were no publication biases in these 4 studies due to the value of P=0.308.
Association between lncRNA HOTAIR and prognosis
OS
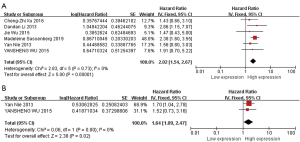
Among the included studies, 6 reported the OS of 856 patients with HNSCC, and this data was pooled for further analysis (26,36,38,42,45,46). The pooled results for the association between high HOTAIR expressions and OS revealed that higher expressions of HOTAIR correlates with worse survival in HNSCC patients (HR =2.02, 95% CI: 1.54–2.67, P<0.00001) (Figure 3A). No heterogeneity between studies was detected (I2=0%, P=0.73).
Through the Begg’s bias test, P=0.707, indicating that there were no publication biases in the 6 pieces of literature (Figure S2A).
DFS
Then we analyzed the pooled HRs of two studies with 236 patients to assess the correlation between high HOTAIR expression and DFS in patients with HNSCC (26,38). Because no heterogeneity (I2=0%, P=0.80), the fixed-effects model was chosen. Compared with the low HOTAIR expression group, the high HOTAIR expression group had a statistically significant reduced DFS (HR =1.64, 95% CI: 1.09–2.47, P=0.02) (Figure 3B).
According to the Begg’s bias test, P=1.000 was obtained, indicating that there were no publication biases in these 2 studies (Figure S2B).
Heterogeneity identification and sensitivity analysis of individual studies
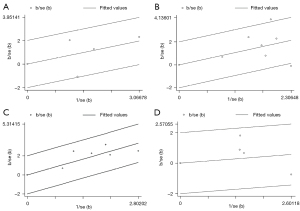
A Galbraith plot of the association between high HOTAIR expression and clinicopathological characteristics, including high T stages, lymphnode metastasis, high TNM stages, and poor histological grades in HNSCC, was constructed to examine the contribution of individual studies to measure heterogeneity and identify outliers (Figure 4). An approximate 95% CI is the area between the two intermittent parallel lines ±2 units above or below the regression line that begins from the origin. Studies outside the CI were identified as outliers.
To make sure the conclusions are credible, we conducted sensitivity analysis, deleting one study at a time for analysis to determine whether any of the studies affected the final pooled results (Figure 5).
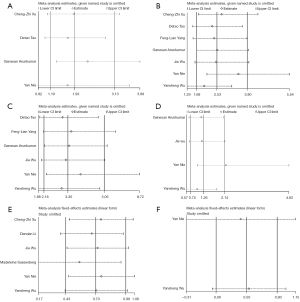
In combination with the aforementioned Galbraith plots and sensitivity analysis, we found the following outliers. Meanwhile, we removed these outliers and conducted a secondary analysis to obtain the new forest plots.
- After removing the study of Ganesan Arunkumar et al. (37), heterogeneity was not detected (I2=0%, P=0.53), and a significant association was found between HOTAIR high expressions and high T stages in HNSCC (OR =2.32, 95% CI: 1.38–3.89, P=0.001) (Figure 6A).
- The study from Yan Nie et al. (38) and Yansheng Wu et al. (26) were deleted to determine the relationship between lymphnode metastasis and HOTAIR expressions. Due to heterogeneity no longer being detected (I2=0%, P=0.72) a fixed-effect model was used to replace the previous random-effects model. The more accurate and reliable correlation results between HOTAIR and lymphnode metastasis can be observed (OR =2.71, 95% CI: 1.57–4.67, P=0.0003) (Figure 6B).
- After deleting the study of Nie et al. (38) relating the association between TNM stages and HOTAIR expressions, the pooled data revealed that the new results (OR =3.92, 95% CI: 2.28–6.72, P<0.00001) and heterogeneity were not detected (I2=0%, P=0.76) (Figure 6C).
- After removing the study of Nie et al. (38), heterogeneity was not detected (I2=0%, P=0.67), a statistically significant association was found between HOTAIR high expressions and poor histological grades in HNSCC (OR =2.21, 95% CI: 1.02–4.83, P=0.046). (Note: The forest plot provided in this paper was generated by Revman software, and the p-value retains two decimal places, but in STATA software, we obtained P=0.05) (Figure 6D).
- The study from Madeleine Sassenberg et al. (46) was removed, then we pooled the results again to reveal the correlation between high HOTAIR expressions and OS in HNSCC patients (HR =1.74, 95% CI: 1.19–2.56, P=0.005), (I2=0%, P=0.81) (Figure 6E).
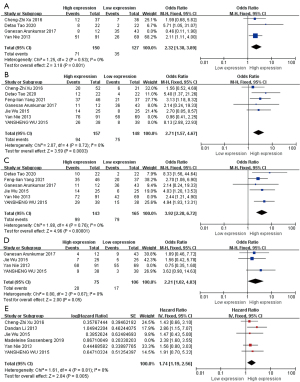
Subgroup analysis
T stages
The four pieces were divided into two groups according to the source of the study population, and meta-analysis was performed respectively. The results are shown in the Figure 7.
Based on the aforementioned subgroup analysis, the overall intergroup heterogeneity was strong, reaching moderate heterogeneity (I2=46%, P=0.13), but in the China group, I2=0%, P=0.53, and there was a significant difference between subgroups (I2=77.3%, P=0.04), suggesting that ethnicity had a significant impact on the meta-analysis results.
The effect size OR of the China group was 2.32 and significant (OR =2.32, 95% CI: 1.38–3.89, P=0.001), implying that there was a significant relationship between high expressions of HOTAIR and high T stages in the China group, and the risk of high T stage was 2.32 times higher in the high expression group than in the low expression group. Second, although there is only one literature on an Indian group, its OR =0.46 (95% CI: 0.11–1.90, P=0.28). This suggests that racial differences may influence HOTAIR expressions in patients with high T stages. Clearly, more studies are needed to further verify this view.
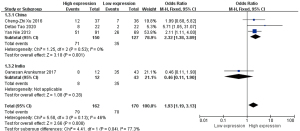
Lymphnode metastasis
The seven studies were divided into three groups based on cut-off differences for HOTAIR gene expressions, and the results were analysed respectively. We divided them into a median group, mean group, and other (Figure 8).
The results from the subgroup analysis showed that the intergroup heterogeneity was significant (I2=50%, P=0.06), and there was a significant difference between subgroups (I2=78.9%, P=0.009), suggesting that the differences in cut-offs had a powerful impact on the meta-analysis results.
The pooled ORs for the cut-offs: median group (OR =5.07, 95% CI: 2.59–9.93, P<0.00001), and the mean group (OR =2.94, 95% CI: 1.20–7.18, P=0.02) support the conclusion that HOTAIR high expressions were significantly related to positive lymph node metastasis. No heterogeneity was found in the two subgroups. However, in the other group, the analysis assessing the association between HOTAIR high expressions and positive lymphnode metastasis showed no statistically significant differences (OR =1.15, 95% CI: 0.59–2.26, P=0.68), although the heterogeneity test showed a reduced heterogeneity (I2=0%, P=0.49).
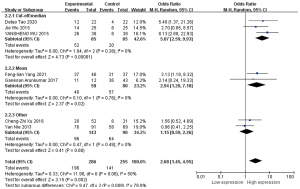
Histological grades
According to different detection methods, the four pieces were divided into a PCR group and ISH group, and meta-analyses were performed respectively (Figure 9).
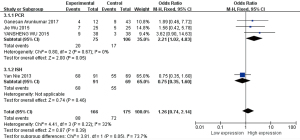
Based on the results from the subgroup analysis, intergroup heterogeneity was present (I2=32%, P=0.22), but after grouping, heterogeneity was not detected (I2=0%, P=0.67) in the PCR group, and a statistically significant association was found between HOTAIR high expressions and poor histological grade in HNSCC (OR =2.21, 95% CI: 1.02–4.83, P=0.05). The risk of poor histological grades was 2.21 times higher in the high expression group than in the low expression group.
The OR of the ISH group was 0.75 (OR =0.75, 95% CI: 0.35–1.60, P=0.46). Although there is only one study in the ISH group, which means nothing for the analysis, the difference between the two groups was significant (I2=77.3%, P=0.04).
Discussion
Identifying potential biomarkers may be crucial in improving prognosis and predicting patient survival outcomes (47,48). Dysregulation of lncRNAs expressions has been found in a variety of cancers, among which HOTAIR is one of the most studied lncRNAs (49,50). The role of lncRNAs in HNSCC has been extensively studied, of course, in addition to HOTAIR, many other lncRNAs play an important role in HNSCC and can be used as biomarkers for predicting tumor stage and prognosis. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) may promote the invasion and metastasis of oral squamous cell carcinoma through EMT process, and its overexpression is related to poor prognosis and significantly shortened OS (51,52). Urothelial carcinoma antigen 1 (UCA1) inhibits the growth and metastasis of oral squamous cell carcinoma cells by activating Wnt/β-catenin, and its overexpression may be related to lymph node metastasis (53). The low expression of maternally expressed gene 3 (MEG3) is associated with lymph node metastasis and TNM stage in patients with HNSCC, and may induce poor prognosis, on the contrary, its overexpression leads to the inhibition of tumor cell proliferation and invasion (54-56).
HOTAIR, as a valuable biomarker reflecting clinical parameters and predicting prognosis, is of great clinical significance in HNSCC. Some studies have shown that HOTAIR expression is positively correlated with tumor volume in oral squamous cell carcinoma (57). Wang et al. found that in advanced T stage or clinical stage, the expression of HOTAIR was significantly higher than that in early stage (58). In previous studies, HOTAIR expression was increased in patients with lymph node metastasis (58,59). Metastasis is one of the main causes of death in patients with HNSCC. Therefore, HOTAIR may be used to predict the survival rate of patients. Clinically, these biomarkers can be used to monitor disease exacerbation, and help to choose treatment methods. In view of this, we conducted the systematic review and meta-analysis. Our objective was to investigate whether HNSCC patients with high expressions of HOTAIR, compared to those with low expressions, had a worse prognosis (OS, DFS) and/or clinicopathological characteristics (tumour T stage, lymphnode metastasis, TNM stage, histological grades).
We performed a meta-analysis of 7 studies involving 546 patients to clarify the relationship between clinicopathologic features and HOTAIR expression in HNSCC patients, and 6 studies comprising 856 HNSCC patients to evaluate the effects of HOTAIR expressions on the prognosis. We found that an accordant effect of high HOTAIR expressions on poor OS and worse clinicopathologic features.
The results indicated that high expressions of HOTAIR was a risk factor for OS in HNSCC (HR =2.02, 95% CI: 1.54–2.67, P<0.00001). In addition, we also found that HOTAIR was significantly correlated with DFS (HR =1.64, 95% CI: 1.09–2.47, P=0.02). These results suggest that lncRNA HOTAIR can be used as a biomarker for head and neck tumor prognosis. However, larger studies are still needed. Only two studies were pooled when assessing the DFS results, resulting in poor reliability. When evaluating OS outcomes, more studies were expected to be included, but because some studies did not provide accurate HR values or Kaplan-Meier curves, and two data from TCGA were excluded to avoid duplication (60,61). In the end, we only included 7 studies, but the study population was indeed much larger than the previous meta-analysis.
Meanwhile, we evaluated the relationships between HOTAIR expressions and HNSCC clinicopathological characteristics. We discovered that high HOTAIR expressions were more likely to lead to high T stages (OR =1.93, 95% CI: 1.19–3.13, P=0.008), lymphnode metastasis (OR =2.68, 95% CI: 1.45–4.95, P=0.002), high TNM stages (OR =3.30, 95% CI: 2.16–5.05, P<0.00001). However, there were no correlations in poor histological grades (OR =1.26, 95% CI: 0.74–2.14, P=0.39).
After deleting a study by heterogeneity identification and sensitivity analysis, fortunately, a statistically significant correlation was found between HOTAIR high expressions and poor histological grades (OR =2.21, 95% CI: 1.02–4.83, P=0.05). However, the deleted literature has the largest weight, and the sample size of the remaining three studies is relatively small, so the number of included studies needs to be increased to generate more reliable results.
The heterogeneity tests and sensitivity analysis of other indicators are as aforementioned. Sensitivity analyses can assess the stability of the results. After the outliers were removed, studies that previously existed heterogeneity are no longer heterogeneous, but in these analyses, after excluding the studies that had a large impact on heterogeneity or sensitivity, there were no significant changes in the pooled results of the remaining studies, only a slight variation in the value of the results, but no qualitative changes, indicating that despite heterogeneity existing, the meaning of the results is relatively stable. The applicability of our results to other ethnic groups may be limited because the majority of the remaining studies were conducted in China.
Then in subgroup analysis, it is worth noting that the elevated HOTAIR levels are highly associated with high T stages in China group, but not in Indian group, suggesting that genetic, ethnic, and environmental factors may influence the progression of squamous cell carcinoma. Previous studies have shown differences in disease risks and prognoses among ethnic groups with head and neck cancer (62,63). But unfortunately, most of our studies were conducted in China, only one from India, which could influence the validity and evidential capability of the results. Hence further large-scale studies should be conducted among different ethnic groups.
The subgroup analysis results of lymphnode metastasis groups confirmed that the differences of gene expression cut-offs had a powerful impact on the results. The conclusion that HOTAIR high expressions were significantly related to positive lymph node metastasis was obtained in the median group and mean group. In the two studies with the staining index (SI) as the cut-off point and the unexplained situations (36,38), pooled results showed no statistically relevant differences. Different cut-off points were associated with high/low gene expression sample sizes, which could be inferred to influence the determinations of the correlations between gene expressions and a clinical feature to some extent. This suggests that in future studies, we should pay attention to the heterogeneity caused by differences in cut-off definitions in single studies, which may affect results.
Among the four studies involving histological grading, three used PCR as the detection method for lncRNA, and only one used ISH. The results of these four studies showed that there were no correlations between high HOTAIR expressions and poor histological grades, but after subgrouping, the pooled results revealed a positive correlation. It may provide thoughts for future research and analysis that different detection methods of gene expression may affect identifying the relationship between HOTAIR expressions and histological grades. ISH can be used to detect the expressions and changes of oncogenes, tumor suppressor genes, and various functional genes at the transcriptional level. Some scholars have studied its sensitivity and specificity (64). As for why the different results were produced in the study from Nie et al. (38), more research data needs to be supplemented to demonstrate whether it is caused by differences in detection methods.
There is much evidence that HOTAIR plays a key role in cancer progression and metastasis. Li et al. showed that HOTAIR promoted the malignant growth of human hepatocellular carcinoma stem cells by inhibiting SETD2 expressions and its phosphorylation (35). Song et al. demonstrated that HOTAIR promoted the development of gastric cancer by regulating and inhibiting the transcription of E-cadherin (28). These studies all showed that high expressions of HOTAIR may activate the characteristics of cancer stem cell (65). Although the mechanisms of tumorigenesis and progression in HNSCC have not been fully elucidated, we verified the negative correlation between high HOTAIR expressions and tumor prognosis by pooled analysis of clinical data.
Conclusions
In conclusion, our meta-analysis confirmed the prognostic role of HOTAIR in HNSCC. High HOTAIR expressions predicted low clinical survival rates and were associated with worse clinicopathologic features. These results revealed that, except for clinical stages and histologic grades of HNSCC patients, HOTAIR may be an excellent biomarker to guide prognosis and therapeutic strategy.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 82073655 and 81773514) and the Scientific Research Level upgrading Project of Anhui Medical University (No. 2020xkjT006).
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-652/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-652/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-652/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Comm S. Italian Cancer Figures - Report 2015 the Burden of Rare Cancers in Italy. Epidemiol Prev 2016;40:7-116. [PubMed]
- Döbrossy L. Epidemiology of head and neck cancer: magnitude of the problem. Cancer Metastasis Rev 2005;24:9-17. [Crossref] [PubMed]
- Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300. [Crossref] [PubMed]
- Arfè A, Malvezzi M, Bertuccio P, et al. Cancer mortality trend analysis in Italy, 1970-2007. Eur J Cancer Prev 2011;20:364-74. [Crossref] [PubMed]
- Marur S, Forastiere AA. Head and Neck Squamous Cell Carcinoma: Update on Epidemiology, Diagnosis, and Treatment. Mayo Clin Proc 2016;91:386-96. [Crossref] [PubMed]
- Al-Jamaei AAH, Helder MN, Forouzanfar T, et al. Age-group-specific trend analyses of oropharyngeal squamous cell carcinoma incidence from 1989 to 2018 and risk factors profile by age-group in 2015-2018: a population-based study in The Netherlands. Eur J Cancer Prev 2022;31:158-65. [PubMed]
- Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18:269-82. [Crossref] [PubMed]
- McDermott JD, Bowles DW. Epidemiology of Head and Neck Squamous Cell Carcinomas: Impact on Staging and Prevention Strategies. Curr Treat Options Oncol 2019;20:43. [Crossref] [PubMed]
- Stordeur S, Schillemans V, Savoye I, et al. Comorbidity in head and neck cancer: Is it associated with therapeutic delay, post-treatment mortality and survival in a population-based study? Oral Oncol 2020;102:104561. [Crossref] [PubMed]
- de França GM, da Silva WR, Medeiros CKS, et al. Five-year survival and prognostic factors for oropharyngeal squamous cell carcinoma: retrospective cohort of a cancer center. Oral Maxillofac Surg 2022;26:261-9. [Crossref] [PubMed]
- Le Campion ACOV, Ribeiro CMB, Luiz RR, et al. Low Survival Rates of Oral and Oropharyngeal Squamous Cell Carcinoma. Int J Dent 2017;2017:5815493. [Crossref] [PubMed]
- Kitamura N, Sento S, Yoshizawa Y, et al. Current Trends and Future Prospects of Molecular Targeted Therapy in Head and Neck Squamous Cell Carcinoma. Int J Mol Sci 2020;22:240. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Dempsey JL, Cui JY. Long Non-Coding RNAs: A Novel Paradigm for Toxicology. Toxicol Sci 2017;155:3-21. [Crossref] [PubMed]
- Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta 2009;1790:936-47. [Crossref] [PubMed]
- Lee H, Zhang Z, Krause HM. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends Genet 2019;35:892-902. [Crossref] [PubMed]
- Yang L, Duff MO, Graveley BR, et al. Genomewide characterization of non-polyadenylated RNAs. Genome Biol 2011;12:R16. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Mahpour A, Mullen AC. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep 2020;3:100177. [Crossref] [PubMed]
- Kunej T, Obsteter J, Pogacar Z, et al. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci 2014;51:344-57. [Crossref] [PubMed]
- Jiang C, Li X, Zhao H, et al. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer 2016;15:62. [Crossref] [PubMed]
- Lin Q, Zhang Y, Liu Y, et al. Effects of long noncoding RNA on prognosis of oral squamous cell carcinoma: A protocol for systematic review and meta-analysis. Medicine (Baltimore) 2021;100:e25507. [Crossref] [PubMed]
- Wang Y, Wang S, Ren Y, et al. The Role of lncRNA Crosstalk in Leading Cancer Metastasis of Head and Neck Squamous Cell Carcinoma. Front Oncol 2020;10:561833. [Crossref] [PubMed]
- Kolenda T, Guglas K, Ryś M, et al. Biological role of long non-coding RNA in head and neck cancers. Rep Pract Oncol Radiother 2017;22:378-88. [Crossref] [PubMed]
- Hu W, Xu W, Shi Y, et al. lncRNA HOTAIR upregulates COX-2 expression to promote invasion and migration of nasopharyngeal carcinoma by interacting with miR-101. Biochem Biophys Res Commun 2018;505:1090-6. [Crossref] [PubMed]
- Wu Y, Zhang L, Zhang L, et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol 2015;46:2586-94. [Crossref] [PubMed]
- Li T, Qin Y, Zhen Z, et al. Long non-coding RNA HOTAIR/microRNA-206 sponge regulates STC2 and further influences cell biological functions in head and neck squamous cell carcinoma. Cell Prolif 2019;52:e12651. [Crossref] [PubMed]
- Song Y, Wang R, Li LW, et al. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int J Oncol 2019;54:77-86. [PubMed]
- Wang Y, Xie Y, Li L, et al. EZH2 RIP-seq Identifies Tissue-specific Long Non-coding RNAs. Curr Gene Ther 2018;18:275-85. [Crossref] [PubMed]
- Rajagopal T, Talluri S, Akshaya RL, et al. HOTAIR LncRNA: A novel oncogenic propellant in human cancer. Clin Chim Acta 2020;503:1-18. [Crossref] [PubMed]
- Özeş AR, Wang Y, Zong X, et al. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci Rep 2017;7:894. [Crossref] [PubMed]
- Deng J, Yang M, Jiang R, et al. Long Non-Coding RNA HOTAIR Regulates the Proliferation, Self-Renewal Capacity, Tumor Formation and Migration of the Cancer Stem-Like Cell (CSC) Subpopulation Enriched from Breast Cancer Cells. PLoS One 2017;12:e0170860. [Crossref] [PubMed]
- Xiao Q, Zheng F, Tang Q, et al. Repression of PDK1- and LncRNA HOTAIR-Mediated EZH2 Gene Expression Contributes to the Enhancement of Atractylenolide 1 and Erlotinib in the Inhibition of Human Lung Cancer Cells. Cell Physiol Biochem 2018;49:1615-32. [Crossref] [PubMed]
- Okugawa Y, Toiyama Y, Hur K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 2014;35:2731-9. [Crossref] [PubMed]
- Li H, An J, Wu M, et al. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 2015;6:27847-64. [Crossref] [PubMed]
- Xu CZ, Jiang C, Wu Q, et al. A Feed-Forward Regulatory Loop between HuR and the Long Noncoding RNA HOTAIR Promotes Head and Neck Squamous Cell Carcinoma Progression and Metastasis. Cell Physiol Biochem 2016;40:1039-51. [Crossref] [PubMed]
- Arunkumar G, Deva Magendhra Rao AK, Manikandan M, et al. Expression profiling of long non-coding RNA identifies linc-RoR as a prognostic biomarker in oral cancer. Tumour Biol 2017;39:1010428317698366. [Crossref] [PubMed]
- Nie Y, Liu X, Qu S, et al. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci 2013;104:458-64. [Crossref] [PubMed]
- Troiano G, Caponio VCA, Boldrup L, et al. Expression of the long non-coding RNA HOTAIR as a prognostic factor in squamous cell carcinoma of the head and neck: a systematic review and meta-analysis. Oncotarget 2017;8:73029-36. [Crossref] [PubMed]
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 2009;339:b2535. [Crossref] [PubMed]
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [Crossref] [PubMed]
- Li D, Feng J, Wu T, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol 2013;182:64-70. [Crossref] [PubMed]
- Tao D, Zhang Z, Liu X, et al. LncRNA HOTAIR promotes the invasion and metastasis of oral squamous cell carcinoma through metastasis-associated gene 2. Mol Carcinog 2020;59:353-64. [Crossref] [PubMed]
- Yang FL, Wei YX, Liao BY, et al. LncRNA HOTAIR regulates the expression of E-cadherin to affect nasopharyngeal carcinoma progression by recruiting histone methylase EZH2 to mediate H3K27 trimethylation. Genomics 2021;113:2276-89. [Crossref] [PubMed]
- Wu J, Xie H. Expression of long noncoding RNA-HOX transcript antisense intergenic RNA in oral squamous cell carcinoma and effect on cell growth. Tumour Biol 2015;36:8573-8. [Crossref] [PubMed]
- Sassenberg M, Droop J, Schulz WA, et al. Upregulation of the long non-coding RNA CASC9 as a biomarker for squamous cell carcinoma. BMC Cancer 2019;19:806. [Crossref] [PubMed]
- M Braden A. Breast cancer biomarkers: risk assessment, diagnosis, prognosis, prediction of treatment efficacy and toxicity, and recurrence. Curr Pharm Des 2014;20:4879-98. [Crossref] [PubMed]
- Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in Hepatocellular Carcinoma: Diagnosis, Prognosis and Treatment Response Assessment. Cells 2020;9:1370. [Crossref] [PubMed]
- Bermúdez M, Aguilar-Medina M, Lizárraga-Verdugo E, et al. LncRNAs as Regulators of Autophagy and Drug Resistance in Colorectal Cancer. Front Oncol 2019;9:1008. [Crossref] [PubMed]
- Deng Q, Sun H, He B, et al. Prognostic value of long non-coding RNA HOTAIR in various cancers. PLoS One 2014;9:e110059. [Crossref] [PubMed]
- Zhou X, Liu S, Cai G, et al. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci Rep 2015;5:15972. [Crossref] [PubMed]
- Fang Z, Zhang S, Wang Y, et al. Long non-coding RNA MALAT-1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins. BMC Cancer 2016;16:706. [Crossref] [PubMed]
- Yang YT, Wang YF, Lai JY, et al. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/β-catenin signaling pathway. Cancer Sci 2016;107:1581-9. [Crossref] [PubMed]
- Chen PY, Hsieh PL, Peng CY, et al. LncRNA MEG3 inhibits self-renewal and invasion abilities of oral cancer stem cells by sponging miR-421. J Formos Med Assoc 2021;120:1137-42. [Crossref] [PubMed]
- Jia LF, Wei SB, Gan YH, et al. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int J Cancer 2014;135:2282-93. [Crossref] [PubMed]
- Zhao YQ, Liu XB, Xu H, et al. MEG3 inhibits cell proliferation, invasion and epithelial-mesenchymal transition in laryngeal squamous cell carcinoma. Eur Rev Med Pharmacol Sci 2019;23:2062-8. [PubMed]
- Vishwakarma S, Pandey R, Singh R, et al. Expression of H19 long non-coding RNA is down-regulated in oral squamous cell carcinoma. J Biosci 2020; [Crossref] [PubMed]
- Wang J, Zhou Y, Lu J, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol 2014;31:148. [Crossref] [PubMed]
- Tang H, Wu Z, Zhang J, et al. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep 2013;7:761-6. [Crossref] [PubMed]
- Lu MY, Liao YW, Chen PY, et al. Targeting LncRNA HOTAIR suppresses cancer stemness and metastasis in oral carcinomas stem cells through modulation of EMT. Oncotarget 2017;8:98542-52. [Crossref] [PubMed]
- Tang Z, Wei G, Zhang L, et al. Signature microRNAs and long noncoding RNAs in laryngeal cancer recurrence identified using a competing endogenous RNA network. Mol Med Rep 2019;19:4806-18. [Crossref] [PubMed]
- Taylor MA, Switchenko J, Stokes W, et al. Incidence trends of squamous cell carcinoma of the head and neck (SCCHN) in the aging population--A SEER-based analysis from 2000 to 2016. Cancer Med 2021;10:6070-7. [Crossref] [PubMed]
- Fakhry C, Westra WH, Wang SJ, et al. The prognostic role of sex, race, and human papillomavirus in oropharyngeal and nonoropharyngeal head and neck squamous cell cancer. Cancer 2017;123:1566-75. [Crossref] [PubMed]
- Suresh K, Shah PV, Coates S, et al. In situ hybridization for high risk HPV E6/E7 mRNA in oropharyngeal squamous cell carcinoma. Am J Otolaryngol 2021;42:102782. [Crossref] [PubMed]
- Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008;133:704-15. [Crossref] [PubMed]


