
C chemokines are prognostic biomarkers correlated with diverse immune cell infiltrations in clear cell renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) is a common malignant tumor of the urinary system, accounting for nearly 90% of all renal cancers, of which clear-cell RCC (ccRCC) is the predominant histologic subtype (1,2). The management of RCC has changed dramatically in the past decades. Antiangiogenic therapy and immunotherapy are the optimal choices for patients with metastatic RCC (3,4). Unfortunately, the development of drug resistance and the low response rate reduce severely the therapeutic effect. To date, the number of robust biomarkers that for the prognosis and immunotherapy response prediction in RCC patients remains scarce (5). Therefore, for a further breakthrough in the treatment of RCC, more attention should be focused on the development of novel predictive biomarkers and therapeutic targets.
Chemokines are a family of cytokines that induce cell migration, especially of leukocytes. Based on the difference of the N-terminal cysteine residues, they are divided into CXC, CC, C, and CX3C subfamilies (6). Through binding to specific transmembrane G-protein-coupled receptors, they regulate the intracellular signaling cascades and participate in various physiological processes (7,8). Due to their versatile functions, numerous studies have been conducted to elucidate the roles of chemokines in cancers (9,10).
The C subfamily consists of X-C motif chemokine ligand 1 (XCL1) and X-C motif chemokine ligand 2 (XCL2), two closely related paralogs (11), that are mainly secreted by natural killer cells (NKs) and activated CD8+ T cells (12,13). XCR1, a member of the G-protein-coupled receptor family, is their common receptor that is selectively expressed on CD8+ dendritic cells (DCs) known for their antigen cross-presentation ability (14). Through binding to XCR1, XCL1 not only exert an important role in the formation of self-tolerance (15), but also initiates cross-presentation of antigens and mediates the cytotoxic immune response (16). Many studies have investigated the fusion of XCL1 and cancer-specific molecules to enhance antigen delivery and CD8+ T-cell response in cancer immunotherapy, also called ‘cancer vaccine’ (17,18). The structure and potency of XCL2 are similar to XCL1 (11). As XCL2 is limited to a small subset of organisms, previous studies mainly focus on XCL1 and ignore to elucidate the biological function of XCL2. Moreover, their functions in cancers are still largely unknown. In this study, using bioinformatics analysis, we comprehensively analyzed the expression, prognostic value, and immune implication of C chemokines in ccRCC. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-424/rc).
Methods
Ethical statement
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Declaration of Helsinki (as revised in 2013).
Tumor IMmune Estimation Resource (TIMER)
TIMER (http://timer.cistrome.org) is a web portal offering the possibility to comprehensively investigate the relationship between diverse immune infiltrations and the gene expression or clinical data across The Cancer Genome Atlas (TCGA) cohorts (19). We analyzed the expression of C chemokines across TCGA cancer types, the correlations between the expression or copy number alteration (CNA) status of C chemokines and diverse immune infiltrations, the correlations between the expression of C chemokines and some immune-related genes, and the prognostic impact of diverse immune infiltrations in kidney renal clear-cell carcinoma (KIRC) dataset. Wilcoxon test was applied for differential expression analysis. Spearman rank correlation test was employed for correlation analysis and the log-rank test for survival analysis. Immune cell infiltration for each CNA category was analyzed by the two-sided Wilcoxon rank-sum test.
Oncomine
Oncomine (http://www.oncomine.org) is a database that integrates the transcriptome data across diverse cancers (20). We investigated the expression of C chemokines in ccRCC. The following screening conditions were set: P value of 0.05, fold change of 2; the gene rank was top 10%, and the data type was mRNA. Student’s t-test was employed for statistical analysis.
The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN)
UALCAN (http://ualcan.path.uab.edu) is an interactive web portal through which the genomics data of TCGA database can be analyzed (21). We explored the expression and promoter methylation levels of C chemokines in KIRC dataset. Student’s t-test was employed for statistical analysis.
Tumor-Immune System Interactions Database (TISIDB)
TISIDB (http://cis.hku.hk/TISIDB) is a web portal that integrates multiple heterogeneous data types that can be utilized for investigating tumor-immune system interactions (22). We performed Spearman correlation analysis of the expression levels of C chemokines and their correlations with the abundance of 28 tumor-infiltrating lymphocyte (TIL) subpopulations, various immunoinhibitors, immunostimulators, MHC molecules, chemokines, and receptors in TCGA KIRC dataset. Spearman rank correlation test was used for statistical analysis.
Quantitative real-time polymerase chain reaction (qPCR)
Human proximal tubule epithelial cell line HK2 and RCC cell lines, including A498, 786-O, SW839, ACHN and Caki1, were purchased from the American Type Culture Collection (ATCC). Total RNA was extracted and reverse-transcribed into cDNA using the PrimeScript RT reagent kit (TaKaRa, Japan). qPCR was performed with the Takara TB Green Premix Ex Taq II (TaKaRa, Japan) using a 7900HT Fast Real-Time PCR System (Applied Biosystems, Japan). The primer sequences were listed as follows: XCL1 forward: 5'-ATGGAAGAGATTCTGGCTAGTGTC-3'; XCL1 reverse: 5'-AGAACCATTACAAGCTGGACTCTAA-3'. XCL2 forward: 5'-ATGATCCAGACCAAGCCAACAG-3'; XCL2 reverse: 5'-GACAGGGTGCCAGAGACTACT-3'. GAPDH forward: 5'-TGAAGGTCGGAGTCAACGGATTTGGT-3'; GAPDH reverse: 5'-CATGTGGGCCATGAGGTCCACCAC-3'. Data were analyzed using the 2-ΔΔCt method. GraphPad Prism 6 software (GraphPad Software, USA) was used for data analysis.
Gene Expression Omnibus (GEO)
GEO is a publicly available genomics database that collects diverse high throughput gene expression data of various samples. We analyzed the expression of C chemokines in three datasets (GSE15641, GSE46699 and GSE53757) that contains the gene expression data of ccRCC tumor tissues and normal kidney tissues. The sample descriptions and gene expression profiles of each microarray study are provided in supplementary materials.
Gene Expression Profiling Interactive Analysis 2 (GEPIA2)
GEPIA2 (http://gepia2.cancer-pku.cn) is a web portal used for investigating gene and gene signature expression across TCGA and Genotype-Tissue Expression (GTEx) cohorts (23). We evaluated the effects of C chemokines on the overall survival (OS) and disease-free survival (DFS) of diverse cancers, including KIRC. Quartile was utilized as a group cutoff value. Hazard ratios (HRs) with 95% confidence intervals and log-rank P values were displayed. Besides, we assessed the correlations between the expression of C chemokines and the genes or gene signatures of interest in KIRC. Spearman rank correlation test was then performed for statistical analysis.
Kaplan-Meier plotter
Kaplan-Meier plotter (http://www.kmplot.com/analysis) is a database used for the assessment of the associations between genes and the survival of 21 cancer types by GEO, EGA, and TCGA data analysis (24). We confirmed the prognostic value of C chemokines in KIRC.
DriverDBv3
DriverDBv3 (http://driverdb.tms.cmu.edu.tw) is an omics database that integrates data on the RNA expression, somatic mutation, CNA, and the methylation and clinical data of driver genes in TCGA cohorts (25). We used it to analyze the expression of C chemokines in KIRC. Kaplan-Meier plots showed the 5-year survival and OS of RCC patients with different expression levels of C chemokines at four survival endpoints, including OS, disease-free interval (DFI), progression-free interval (PFI), and disease-specific survival (DSS). The mean was used as the cutoff value. HRs and log-rank P values were provided. Moreover, based on the HRs of C chemokines and their related genes, we constructed the survival networks to illustrate their synergistic effects on patient survival. For two genes, when the HR value was 1.5-fold greater than that of each of them, they were considered to work synergistically. The positive direction of HR was then presented.
LinkedOmics
LinkedOmics (http://www.linkedomics.org) is a database containing multi-omics and clinical data of different TCGA cohorts (26). Using Spearman test, we investigated the genes that were positively or negatively correlated (P value <0.05 and false discovery rate <0.01) with the expression of C chemokines in KIRC.
GeneMANIA
GeneMANIA (http://www.genemania.org) is a web portal used for predicting the related genes of a given gene or a gene list by the assessment of available genomics and proteomics data (27). We constructed the protein-protein interaction (PPI) networks of C chemokines and their related genes. The networks were composed of physical interactions, co-expression, predicted, co-localization, pathway, genetic interactions, and shared protein domains, and were then represented as different colored lines. The involved functions of these genes are listed and represented in different color chips.
The Database for Annotation, Visualization and Integrated Discovery (DAVID)
DAVID (https://david.ncifcrf.gov) is an integrated biological database that allows the systematic analysis of the biological significance of given gene lists (28). We performed Gene Ontology (GO) annotation of the gene list derived from GeneMANIA in three categories, including biological process (BP), molecular function (MF) and cellular component (CC). Besides, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed also. The results obtained are displayed in bubble diagrams.
Results
C chemokines are upregulated in ccRCC
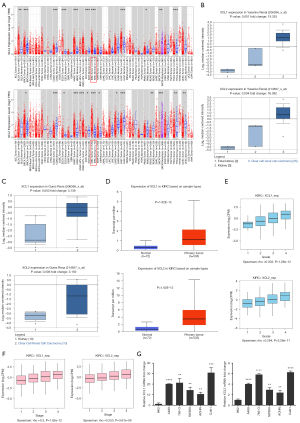
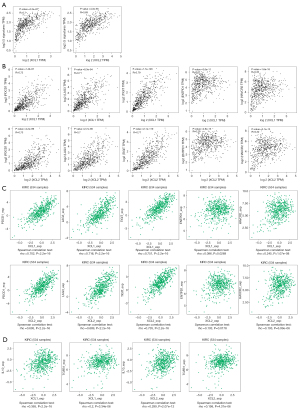
To investigate the transcriptional levels of C chemokines in human cancers, we detected the differential expression of C chemokines across TCGA cohorts using TIMER. Our results indicated that both XCL1 and XCL2 were upregulated in multiple tumor tissues, including KIRC (Figure 1A). Then, we performed a search in the Oncomine database and found the upregulation of XCL1 and XCL2 in ccRCC tumor tissues as compared with that in normal kidney tissues in two datasets, including Yusenko Renal (Figure 1B) and Gumz Renal (Figure 1C). UALCAN further revealed the overexpression of XCL1 and XCL2 in ccRCC primary tumor tissues as compared with their expression in normal tissues (Figure 1D). Besides, XCL1 and XCL2 showed pathological grade and clinical stage specific expression as we observed their significantly increased levels following the progression of the tumor grade and stage (Figure 1E,1F). Furthermore, we detected the expression of XCL1 and XCL2 in RCC cell lines by qPCR. Results suggested that both XCL1 and XCL2 were overexpressed in RCC cell lines compared with that in HK2 cells (Figure 1G). We also found the upregulation of XCL1 in ccRCC tumor tissues in three GEO datasets (GSE15641, GSE46699 and GSE53757) (Figure S1). In summary, C chemokines may participate in the initiation and progression of ccRCC.
Promoter hypomethylation contributes to the overexpression of C chemokines in ccRCC
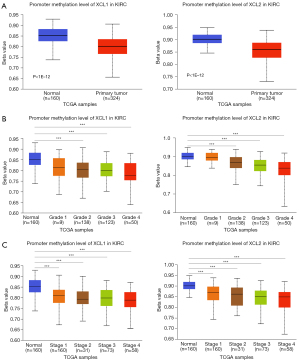
To investigate the potential influence factors for the overexpression of C chemokines in ccRCC, the promoter methylation profiles of C chemokines in KIRC were analyzed by UALCAN using sample types, tumor grades, and stages as criteria. As can be seen in Figure 2A, the promoters of XCL1 and XCL2 were in a hypomethylation status in ccRCC tumor tissues. Moreover, the methylation of the XCL1 and XCL2 promoters decreased with the increase in the tumor grade and stage (Figure 2B,2C). Therefore, promoter hypomethylation may contribute to the upregulation of C chemokines in ccRCC.
The excessive expression of C chemokines predicts poor prognosis in ccRCC
The prognostic value of C chemokines was evaluated through analysis of the clinical data in TCGA database using GEPIA2. The impacts of C chemokines on OS and DFS in diverse cancer types are illustrated in heat maps. We established that the expression of XCL1 and XCL2 in several cancers was significantly associated with OS, rather than with DFS (Figure 3A,3B). KIRC patients with higher levels of XCL1 and XCL2 had significantly poorer OS (Figure 3A), whereas no difference was observed in DFS (Figure 3B). Similar results were obtained from the Kaplan-Meier plotter (Figure S2). The associations between C chemokines and the OS of KIRC with different clinical features are presented in Table 1.
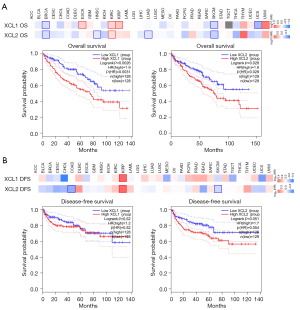
Table 1
| Clinical features | XCL1 overall survival (n=530) | XCL2 overall survival (n=530) | |||||
|---|---|---|---|---|---|---|---|
| n | HR (95% CI) | P | n | HR (95% CI) | P | ||
| Gender | |||||||
| Female | 186 | 2.07 (1.22–3.49) | 0.006* | 186 | 1.85 (1.11–3.07) | 0.016* | |
| Male | 344 | 1.69 (1.16–2.46) | 0.005* | 344 | 2.04 (1.22–3.42) | 0.006* | |
| Stage | |||||||
| 1 | 265 | 1.69 (0.91–3.16) | 0.095 | 265 | 2.27 (1.22–4.24) | 0.008* | |
| 2 | 57 | 0.53 (0.16–1.73) | 0.280 | 57 | 3.23 (0.71–14.61) | 0.110 | |
| 3 | 123 | 0.62 (0.35–1.10) | 0.100 | 123 | 0.59 (0.33–1.05) | 0.072 | |
| 4 | 82 | 1.97 (1.03–3.79) | 0.038* | 82 | 1.44 (0.85–2.42) | 0.170 | |
| Grade | |||||||
| 1 | 14 | – | – | 14 | – | – | |
| 2 | 227 | 0.67 (0.36–1.27) | 0.220 | 227 | 2.23 (1.18–4.20) | 0.011* | |
| 3 | 206 | 2.00 (1.24–3.22) | 0.004* | 206 | 1.61 (1.02–2.56) | 0.041* | |
| 4 | 75 | 0.56 (0.29–1.10) | 0.087 | 75 | 0.66 (0.37–1.18) | 0.160 | |
| Mutation burden | |||||||
| High | 168 | 1.44 (0.81–2.57) | 0.210 | 168 | 1.55 (0.89–2.70) | 0.120 | |
| Low | 164 | 2.26 (1.02–5.00) | 0.038* | 164 | 2.11 (0.94–4.71) | 0.063 | |
*, significant difference. KIRC, kidney renal clear-cell carcinoma; HR, hazard ratio.
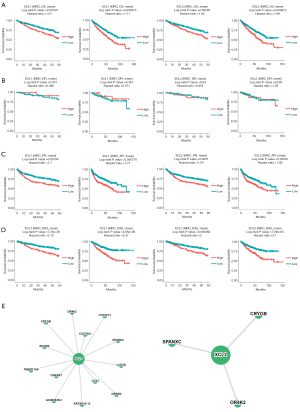
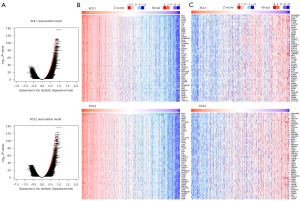
In addition to the analyses performed using GEPIA2 and the Kaplan-Meier plotter, we further assessed the prognostic value of C chemokines using DriverDBv3. Kaplan-Meier plots depict the survival of KIRC patients at four survival endpoints, including OS (Figure 4A), DFI (Figure 4B), PFI (Figure 4C) and DSS (Figure 4D). As can be observed in the figure, the high levels of XCL1 and XCL2 predicted poor OS, PFI, and DSS for both the 5-year survival and the OS of KIRC patients. Besides, we constructed the survival networks of C chemokines and their related genes, the high expression of which synergistically with C chemokines predicted poor survival of cancer patients (Figure 4E). The synergistic effects of each gene are specified in Figure S3. Altogether, the expression of C chemokines can be used for prognosis prediction in ccRCC patients.
Functional analysis of C chemokines in ccRCC
We investigated the impact of C chemokines on the transcriptome of ccRCC via LinkedOmics. As visible in Figure 5A, a total number of 20,159 genes from 533 KIRC samples were analyzed, 6,007 of which were positively correlated with XCL1 expression, whereas 4,148 were negatively correlated with XCL1 expression. For XCL2, we detected 6,360 positively correlated genes and 4,154 negatively correlated genes. The heat maps illustrate the top 50 genes that are positively (Figure 5B) and negatively (Figure 5C) correlated with XCL1 and XCL2, respectively.
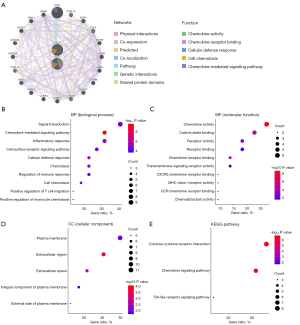
To investigate the functions of C chemokines, PPI networks were constructed using GeneMANIA. As illustrated in Figure 6A, 20 genes were verified to be closely associated with C chemokines. Then, DAVID was employed to perform GO and KEGG analyses of these genes. The top 10 most highly enriched BP items are depicted in Figure 6B, such as the signal transduction, chemokine-mediated signaling pathway, inflammatory response, cell surface receptor signaling pathway, and the cellular defense response. Concerning the MF category, chemokine activity, carbohydrate binding, receptor activity, receptor binding, and chemokine receptor binding were enriched (Figure 6C). Besides, plasma membrane, extracellular region, extracellular space, integral component of plasma membrane, and external side of plasma membrane were enriched in CC ontology (Figure 6D). KEGG analysis revealed that these genes were involved mainly in the cytokine-cytokine receptor interaction, chemokine signaling pathway, and the Toll-like receptor signaling pathway (Figure 6E).
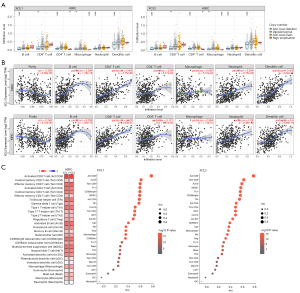
C chemokines are associated with the infiltration of diverse immune cells in ccRCC
Considering the vital role of immune cells in the initiation, progression, and treatment of cancers, we investigated C chemokines-related immune infiltration variations in ccRCC. First, we analyzed the abundances of immune cells in different CNA statuses of C chemokines in RCC tumor tissues. Our results showed that the arm-level deletion was significantly associated with the infiltration levels of several immune cell subpopulations (Figure 7A). Moreover, the expression levels of C chemokines were positively correlated with B-cell, CD8+ T-cell, neutrophil, and DC infiltration (Figure 7B). The results obtained from TISIDB further revealed that C chemokines levels were positively correlated with the abundance of 25 types of immune cells in RCC tumor tissues, including various subtypes of CD8+ T cells, CD4+ T cells, helper T cells (Th), B cells, DC, NK, natural killer T cells (NKT), gamma delta T cells (Tgd), mast cells, monocytes, macrophages, myeloid-derived suppressor cells (MDSC), and regulatory T cells (Treg) (Figure 7C).
We also analyzed the expression of diverse immune cell markers in KIRC using TIMER. The results we obtained revealed a significant positive correlation between C chemokines and several cell types with or without adjustment for tumor purity, including T cells, CD8+ T cells, B cells, monocytes, tumor-associated macrophages (TAMs), M2 macrophages, neutrophils, and NK and Th1 cells (Table 2). The positive correlation between C chemokines and the monocyte, TAM and M2 macrophage recruitment was also validated by the GEPIA2 analysis results (Table 3), which indicated the association between C chemokines and macrophage polarization in RCC.
Table 2
| Description | Cell markers | XCL1 | XCL2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | |||||||||
| R | P | R | P | R | P | R | P | |||||
| T cell | CD3D | 0.748 | **** | 0.728 | **** | 0.762 | **** | 0.736 | **** | |||
| CD3E | 0.745 | **** | 0.726 | **** | 0.764 | **** | 0.742 | **** | ||||
| CD2 | 0.742 | **** | 0.723 | **** | 0.755 | **** | 0.730 | **** | ||||
| CD8+ T cell | CD8A | 0.710 | **** | 0.686 | **** | 0.707 | **** | 0.672 | **** | |||
| CD8B | 0.697 | **** | 0.676 | **** | 0.698 | **** | 0.668 | **** | ||||
| B cell | CD19 | 0.463 | **** | 0.426 | **** | 0.471 | **** | 0.432 | **** | |||
| CD79A | 0.529 | **** | 0.499 | **** | 0.542 | **** | 0.504 | **** | ||||
| Monocyte | CD86 | 0.442 | **** | 0.395 | **** | 0.418 | **** | 0.364 | **** | |||
| CSF1R | 0.323 | **** | 0.269 | **** | 0.325 | **** | 0.265 | **** | ||||
| TAM | CCL2 | 0.054 | 0.211 | −0.012 | 0.790 | 0.038 | 0.385 | −0.025 | 0.598 | |||
| CD68 | 0.316 | **** | 0.294 | **** | 0.288 | **** | 0.256 | **** | ||||
| IL10 | 0.344 | **** | 0.277 | **** | 0.292 | **** | 0.211 | **** | ||||
| M1 Macrophage | NOS2 | −0.149 | *** | −0.197 | **** | −0.104 | * | −0.155 | *** | |||
| IRF5 | 0.323 | **** | 0.295 | **** | 0.308 | **** | 0.282 | **** | ||||
| PTGS2 | −0.076 | 0.080 | −0.113 | * | −0.095 | * | −0.133 | ** | ||||
| M2 macrophage | CD163 | 0.155 | *** | 0.109 | * | 0.141 | ** | 0.085 | 0.069 | |||
| VSIG4 | 0.263 | **** | 0.204 | **** | 0.261 | **** | 0.189 | **** | ||||
| MS4A4A | 0.268 | **** | 0.210 | **** | 0.248 | **** | 0.183 | **** | ||||
| Neutrophil | CEACAM8 | −0.083 | 0.056 | −0.069 | 0.140 | −0.069 | 0.114 | −0.059 | 0.203 | |||
| ITGAM | 0.289 | **** | 0.239 | **** | 0.267 | **** | 0.206 | **** | ||||
| CCR7 | 0.431 | **** | 0.378 | **** | 0.438 | **** | 0.393 | **** | ||||
| NK | KIR2DL1 | 0.153 | *** | 0.160 | *** | 0.227 | **** | 0.229 | **** | |||
| KIR2DL3 | 0.160 | *** | 0.180 | *** | 0.270 | **** | 0.278 | **** | ||||
| KIR2DL4 | 0.452 | **** | 0.424 | **** | 0.486 | **** | 0.473 | **** | ||||
| KIR3DL1 | 0.146 | *** | 0.167 | *** | 0.228 | **** | 0.245 | **** | ||||
| KIR3DL2 | 0.276 | **** | 0.271 | **** | 0.377 | **** | 0.378 | **** | ||||
| KIR3DL3 | 0.168 | **** | 0.138 | ** | 0.184 | **** | 0.171 | *** | ||||
| KIR2DS4 | 0.136 | ** | 0.145 | ** | 0.204 | **** | 0.209 | **** | ||||
| DC | HLA-DPB1 | 0.532 | **** | 0.514 | **** | 0.531 | **** | 0.513 | **** | |||
| HLA-DQB1 | 0.396 | **** | 0.355 | **** | 0.398 | **** | 0.360 | **** | ||||
| HLA-DRA | 0.511 | **** | 0.495 | **** | 0.495 | **** | 0.475 | **** | ||||
| HLA-DPA1 | 0.534 | **** | 0.514 | **** | 0.509 | **** | 0.485 | **** | ||||
| CD1C | 0.181 | **** | 0.131 | ** | 0.194 | **** | 0.145 | ** | ||||
| NRP1 | −0.179 | **** | −0.224 | **** | −0.139 | ** | −0.186 | **** | ||||
| ITGAX | 0.323 | **** | 0.297 | **** | 0.312 | **** | 0.271 | **** | ||||
| Th1 | TBX21 | 0.398 | **** | 0.366 | **** | 0.545 | **** | 0.519 | **** | |||
| STAT4 | 0.457 | **** | 0.408 | **** | 0.513 | **** | 0.471 | **** | ||||
| STAT1 | 0.454 | **** | 0.417 | **** | 0.406 | **** | 0.353 | **** | ||||
| IFNG | 0.716 | **** | 0.699 | **** | 0.716 | **** | 0.693 | **** | ||||
| TNF | 0.254 | **** | 0.212 | **** | 0.262 | **** | 0.226 | **** | ||||
| Th2 | GATA3 | 0.294 | **** | 0.272 | **** | 0.32 | **** | 0.313 | **** | |||
| STAT6 | −0.108 | * | −0.106 | * | −0.071 | 0.100 | −0.068 | 0.144 | ||||
| STAT5A | 0.379 | **** | 0.324 | **** | 0.344 | **** | 0.290 | **** | ||||
| IL13 | 0.084 | 0.052 | 0.040 | 0.397 | 0.120 | ** | 0.101 | * | ||||
| Tfh | BCL6 | −0.128 | ** | −0.143 | ** | −0.079 | 0.067 | −0.091 | * | |||
| IL21 | 0.162 | *** | 0.135 | ** | 0.226 | **** | 0.206 | **** | ||||
| Th17 | STAT3 | −0.081 | 0.062 | −0.130 | ** | −0.109 | * | −0.156 | *** | |||
| IL17A | 0.085 | 0.051 | 0.046 | 0.326 | 0.072 | 0.099 | 0.036 | 0.437 | ||||
| Treg | FOXP3 | 0.646 | **** | 0.594 | **** | 0.632 | **** | 0.596 | **** | |||
| CCR8 | 0.563 | **** | 0.511 | **** | 0.546 | **** | 0.501 | **** | ||||
| STAT5B | −0.194 | **** | −0.206 | **** | −0.162 | *** | −0.172 | *** | ||||
| TGFB1 | 0.092 | * | 0.037 | 0.431 | 0.091 | * | 0.060 | 0.202 | ||||
R, R-value of Spearman’s correlation. None, correlation without adjustment. Purity, correlation adjusted by tumor purity. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. KIRC, kidney renal clear-cell carcinoma; TIMER, Tumor IMmune Estimation Resource; TAM, tumor-associated macrophage; NK, natural killer cell; DC, dendritic cell; Th1, Type 1 T helper cell; Th2, Type 2 T helper cell; Tfh, T follicular helper cell; Th17, Type 17 T helper cell; Treg, regulatory T cell.
Table 3
| Description | Cell markers and signatures | XCL1 | XCL2 | |||
|---|---|---|---|---|---|---|
| R | P | R | P | |||
| Monocyte | CD86 | 0.530 | **** | 0.510 | **** | |
| CSF1R | 0.450 | **** | 0.450 | **** | ||
| Signature | 0.500 | **** | 0.490 | **** | ||
| TAM | CCL2 | 0.092 | * | 0.075 | 0.069 | |
| CD68 | 0.410 | **** | 0.400 | **** | ||
| IL10 | 0.390 | **** | 0.350 | **** | ||
| Signature | 0.350 | **** | 0.330 | **** | ||
| M1 macrophage | NOS2 | −0.064 | 0.120 | −0.024 | 0.56 | |
| IRF5 | 0.270 | **** | 0.250 | **** | ||
| PTGS2 | −0.180 | **** | −0.210 | **** | ||
| Signature | −0.037 | 0.360 | −0.044 | 0.280 | ||
| M2 macrophage | CD163 | 0.380 | **** | 0.380 | **** | |
| VSIG4 | 0.380 | **** | 0.400 | **** | ||
| MS4A4A | 0.410 | **** | 0.400 | **** | ||
| Signature | 0.410 | **** | 0.410 | **** | ||
R, R-value of Spearman’s correlation. Signature, the combination of immune cell markers. *, P<0.05; ****, P<0.0001. TAM, tumor-associated macrophage; KIRC, kidney renal clear-cell carcinoma; GEPIA2, Gene Expression Profiling Interactive Analysis 2.
Further, we explored the prognosis relevance of tumor-infiltrating immune cells in ccRCC. According to the results obtained after the searches in several databases, KIRC patients with lower abundance of CD4+ T cells and DCs had poorer survival, whereas higher levels of monocyte, macrophage, Tfh, MDSC, Treg, and NKT infiltration predicted poorer survival (Figure S4). To sum up, C chemokines influence the composition of the immune microenvironment by the recruitment of diverse immune cells, which may regulate ccRCC development and prognosis.
C chemokines are associated with T-cell exhaustion in ccRCC
T-cell exhaustion is an important mechanism of tumor immune escape, which seriously affects the therapeutic efficacy of immunotherapy. Thus, we subsequently investigated the implication of C chemokines in T-cell exhaustion. As exhausted T cells can be characterized by the persistent overexpression of multiple inhibitory receptors (IRs) (29), we performed correlation analysis between the expression of C chemokines and a gene signature that comprises several common IRs (30), including ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1) (31), hepatitis A virus cellular receptor 2 (HAVCR2) (32), lymphocyte activating 3 (LAG3) (33), programmed cell death 1 (PDCD1) (34), and T cell immunoreceptor with Ig and ITIM domains (TIGIT) (35). As can be observed in Figure 8A,8B, both XCL1 and XCL2 had a positive correlation with the signature as well as with each gene of the signature in KIRC. Consistently, similar results were obtained in TISIDB (Figure 8C). In addition, we found that XCL1 and XCL2 expression correlated more significantly with PDCD1, LAG3 and TIGIT expression than other checkpoint molecules, which indicated the importance of PDCD1, LAG3 and TIGIT in the signature.
A previous study showed the implication of IL10 and TGF-β in the induction of T-cell exhaustion (29). Here, we found that C chemokines were positively correlated with the expression of IL10 and TGF-β in KIRC (Figure 8D and Figure S5). In summary, C chemokines are positively correlated with the expression of several inhibitory receptors and cytokines that are associated with T-cell exhaustion in ccRCC. Their implication in the development of T-cell exhaustion needs further research in the future.
C chemokines are positively correlated with the expression of several immune checkpoints in ccRCC
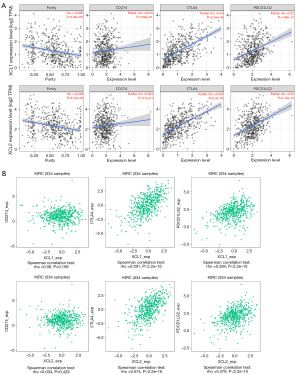
Currently, many immune checkpoint inhibitors (ICIs) targeting programmed cell death 1 (PD1, PDCD1), programmed death ligand 1 (PD-L1, CD274), PD-L2 (PDCD1LG2), and cytotoxic T lymphocyte-associated antigen 4 (CTLA4) have been applied for cancer treatment. To investigate the influence of C chemokines on immunotherapy, we investigated the correlations between C chemokines and immune checkpoints. Figure 8B,8C illustrate the positive correlation between C chemokines and PD1 expression. Using TIMER and TISIDB, we further obtained similar results for PD-L2 and CTLA4 (Figure 9). In addition, other chemokines, receptors, and immunomodulators, including immunoinhibitors, immunostimulators, and major histocompatibility complex (MHC) molecules, which were associated with C chemokines expression, are displayed in Figure S6. In summary, C chemokines are positively correlated with the expression of several immune checkpoints, which may contribute to the prediction of immunotherapy response in ccRCC.
Discussion
Chemokines modulate the composition of tumor microenvironment (TME) and influence the development of cancers and the therapeutic outcomes by inducing chemotaxis of diverse cells, particularly immune cells. C chemokines are a subfamily of chemokines that have been confirmed to participate in innate and adaptive immunity; however, their involvement in cancers has not been well elucidated.
A previous study showed that the expression of XCL1, XCL2, and XCR1 in epithelial ovarian carcinoma tissues and cell lines, and exogenous XCL1 and XCL2 facilitated the proliferation and migration of XCR1-positive cells (36). In lung cancer, XCL1 promoted the proliferation and migration of cancer cells through the JAK2/STAT3 signaling pathway (37). Additionally, in breast cancer, XCL1 stimulated metastasis via enhancing epithelial-mesenchymal transition (EMT) (38). Moreover, XCL1 is a potential biomarker that can be used to predict the malignant transformation of mature cystic teratoma into squamous cell carcinoma (39). XCL2 was found to be upregulated in lung cancer tumor tissues and increased with higher degree of tumor malignancy (40). These results indicate the potential pro-tumor impact of C chemokines. In our study, both XCL1 and XCL2 were overexpressed in ccRCC tumor tissues and cell lines and had a tumor grade and stage specific expression, suggesting their implication in the initiation and progression of ccRCC. Promoter methylation is a crucial epigenetic process that induces gene silencing in cancer (41). We detected the hypomethylation status of XCL1 and XCL2 promoters in ccRCC tumor tissues, which was a potential factor for the upregulation of C chemokines in ccRCC. Furthermore, survival analysis revealed that high levels of XCL1 and XCL2 predicted poor OS, PFI, and DSS of ccRCC patients, which highlighted the prognostic value of XCL1 and XCL2 in ccRCC.
Function analysis of C chemokines-related genes showed the enrichment of cytokine-cytokine receptor interaction, chemokine signaling pathway, and Toll-like receptor (TLR) signaling pathway. They are important regulators of various biological processes, such as innate and adaptive immunity and inflammation, and play a crucial role in the development of cancers (7,10). Accumulating evidence has confirmed the pro-metastatic effect of the XCL1/XCR1 axis (36-38). Moreover, many studies have focused on applying small-molecule modulators targeting TLRs for cancer treatment (42,43). For example, the combination of immune modulatory oligonucleotide, an agonist of TLR9, and everolimus synergistically inhibited the growth and angiogenesis of RCC (44). Therefore, C chemokines are potential future therapeutic targets of RCC.
In recent years, with the introduction of the immune checkpoint blockade (ICB), the therapy landscape of several solid tumors including RCC has been substantially altered (45). Although ICB has exerted remarkable therapeutic effects for cancer treatment, only approximately 20% of the patients acquire sustained benefits (46). Recent evidence has revealed that the dysfunction and abnormal distribution of immune cell subsets in TME seriously affect the efficacy of immunotherapy (47). Therefore, investigating immune cell infiltration in TME is conducive to ascertaining the immune status and choosing appropriate therapeutic strategies.
Our research established that C chemokines were associated with the recruitment of immune cells that participate in either antitumor immunity, such as CD8+ T cell, CD4+ T cell, DC and NK cell, or immunosuppression, such as MDSC and Treg (48) in ccRCC. Furthermore, the positive correlation of C chemokines with the abundance of monocytes, TAMs, and M2 macrophages indicated the modulation of C chemokines on macrophage polarization in ccRCC. Previous studies suggested that the abundance of tumor-infiltrating immune cells affects the survival of cancer patients (49-51). We observed that high levels of tumor-infiltrating CD4+ T cells, and DCs predicted long OS, whereas those of monocytes, macrophages, Tfhs, MDSCs, Tregs, and NKTs predicted short OS in ccRCC. Overall, C chemokines may affect the prognosis of ccRCC through regulating the infiltration of various immune cells.
In patients with chronic infections and cancers, CD8+ T cells gradually become dysfunctional due to their persistent exposure to the stimulation of tumor antigens and inflammation, which is called T-cell exhaustion. Due to their reduced ability to secrete cytokines and the overexpression of IRs, exhausted T cells are in a functionally suppressed state and do not perform well in antitumor immunity, which is one of the major factors of tumor immune evasion (52,53). Further research on the development of T-cell exhaustion would provide a promising strategy for the improvement of immunotherapy outcomes and for the exploration of novel therapeutic targets. In this study, we observed a positive correlation of C chemokines and the signature of T-cell exhaustion in ccRCC. Besides, the levels of C chemokines were positively correlated with the expression of IL10 and TGF-β which induce T-cell exhaustion (54,55). From an immunotherapy point, XCL1 expression was found to be associated with increased PD-L1 expression (39). In ccRCC, we detected high levels of C chemokines which were accompanied by increased PD-1, PD-L2, and CTLA4. In summary, C chemokines are positively correlated with the expression of several inhibitory receptors and cytokines that are associated with T-cell exhaustion and several immune checkpoints in ccRCC, which may help us to understand the immune status in TME and contribute to the prediction of immunotherapy response in ccRCC. Considering the correlation between C chemokines expression and diverse immune cell infiltration, this may affect the composition of the tumor immune microenvironment and regulate immune response. Therefore, more research should be performed to illustrate the function of C chemokines in RCC in the future.
Our study has some limitations. We could not comprehensively understand the immune infiltration status through analyzing the transcriptional levels of immune-related genes. The results need to be validated in other cohorts and basic research. In summary, our research provides preliminary insights into the prognostic value and immune implication of C chemokines in ccRCC, which is conducive to the prediction of survival and immunotherapy response and the development of novel therapeutic targets for ccRCC.
Acknowledgments
We express our gratitude to TCGA, TIMER, Oncomine, UALCAN, TISIDB, GEO, GEPIA2, Kaplan-Meier plotter, DriverDBv3, LinkedOmics, GeneMANIA, and DAVID for providing their platforms and to the contributors for uploading their valuable datasets.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-424/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-424/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer 2018;103:356-87. [Crossref] [PubMed]
- Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers 2017;3:17009. [Crossref] [PubMed]
- Escudier B, Porta C, Schmidinger M, et al. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol 2019;30:706-20. [Crossref] [PubMed]
- Barata PC, Rini BI. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J Clin 2017;67:507-24. [Crossref] [PubMed]
- Raimondi A, Sepe P, Zattarin E, et al. Predictive Biomarkers of Response to Immunotherapy in Metastatic Renal Cell Cancer. Front Oncol 2020;10:1644. [Crossref] [PubMed]
- Hughes CE, Nibbs RJB. A guide to chemokines and their receptors. FEBS J 2018;285:2944-71. [Crossref] [PubMed]
- Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med 2006;354:610-21. [Crossref] [PubMed]
- Le Y, Zhou Y, Iribarren P, et al. Chemokines and chemokine receptors: their manifold roles in homeostasis and disease. Cell Mol Immunol 2004;1:95-104. [PubMed]
- Do HTT, Lee CH, Cho J. Chemokines and their Receptors: Multifaceted Roles in Cancer Progression and Potential Value as Cancer Prognostic Markers. Cancers (Basel) 2020;12:287. [Crossref] [PubMed]
- Vilgelm AE, Richmond A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front Immunol 2019;10:333. [Crossref] [PubMed]
- Fox JC, Nakayama T, Tyler RC, et al. Structural and agonist properties of XCL2, the other member of the C-chemokine subfamily. Cytokine 2015;71:302-11. [Crossref] [PubMed]
- Stievano L, Piovan E, Amadori A. C and CX3C chemokines: cell sources and physiopathological implications. Crit Rev Immunol 2004;24:205-28. [Crossref] [PubMed]
- Lei Y, Takahama Y. XCL1 and XCR1 in the immune system. Microbes Infect 2012;14:262-7. [Crossref] [PubMed]
- Crozat K, Guiton R, Contreras V, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp Med 2010;207:1283-92. [Crossref] [PubMed]
- Lei Y, Ripen AM, Ishimaru N, et al. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med 2011;208:383-94. [Crossref] [PubMed]
- Dorner BG, Dorner MB, Zhou X, et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 2009;31:823-33. [Crossref] [PubMed]
- Mizumoto Y, Hemmi H, Katsuda M, et al. Anticancer effects of chemokine-directed antigen delivery to a cross-presenting dendritic cell subset with immune checkpoint blockade. Br J Cancer 2020;122:1185-93. [Crossref] [PubMed]
- Botelho NK, Tschumi BO, Hubbell JA, et al. Combination of Synthetic Long Peptides and XCL1 Fusion Proteins Results in Superior Tumor Control. Front Immunol 2019;10:294. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep 2021;11:6047. [Crossref] [PubMed]
- Liu SH, Shen PC, Chen CY, et al. DriverDBv3: a multi-omics database for cancer driver gene research. Nucleic Acids Res 2020;48:D863-70. [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Blank CU, Haining WN, Held W, et al. Defining 'T cell exhaustion'. Nat Rev Immunol 2019;19:665-74. [Crossref] [PubMed]
- Liu Y, Zhou N, Zhou L, et al. IL-2 regulates tumor-reactive CD8+ T cell exhaustion by activating the aryl hydrocarbon receptor. Nat Immunol 2021;22:358-69. [Crossref] [PubMed]
- Canale FP, Ramello MC, Núñez N, et al. CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8+ T Cells. Cancer Res 2018;78:115-28. [Crossref] [PubMed]
- Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol 2020;20:173-85. [Crossref] [PubMed]
- Andrews LP, Marciscano AE, Drake CG, et al. LAG3 (CD223) as a cancer immunotherapy target. Immunol Rev 2017;276:80-96. [Crossref] [PubMed]
- Barber DL, Wherry EJ, Masopust D, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006;439:682-7. [Crossref] [PubMed]
- Johnston RJ, Comps-Agrar L, Hackney J, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell 2014;26:923-37. [Crossref] [PubMed]
- Kim M, Rooper L, Xie J, et al. The lymphotactin receptor is expressed in epithelial ovarian carcinoma and contributes to cell migration and proliferation. Mol Cancer Res 2012;10:1419-29. [Crossref] [PubMed]
- Wang T, Han S, Wu Z, et al. XCR1 promotes cell growth and migration and is correlated with bone metastasis in non-small cell lung cancer. Biochem Biophys Res Commun 2015;464:635-41. [Crossref] [PubMed]
- Do HTT, Cho J. Involvement of the ERK/HIF-1α/EMT Pathway in XCL1-Induced Migration of MDA-MB-231 and SK-BR-3 Breast Cancer Cells. Int J Mol Sci 2020;22:89. [Crossref] [PubMed]
- Tamura R, Yoshihara K, Nakaoka H, et al. XCL1 expression correlates with CD8-positive T cells infiltration and PD-L1 expression in squamous cell carcinoma arising from mature cystic teratoma of the ovary. Oncogene 2020;39:3541-54. [Crossref] [PubMed]
- Zhou B, Xu H, Ni K, et al. Expression of Chemokine XCL2 and CX3CL1 in Lung Cancer. Med Sci Monit 2016;22:1560-5. [Crossref] [PubMed]
- Morgan AE, Davies TJ, Mc Auley MT. The role of DNA methylation in ageing and cancer. Proc Nutr Soc 2018;77:412-22. [Crossref] [PubMed]
- Wang Y, Zhang S, Li H, et al. Small-Molecule Modulators of Toll-like Receptors. Acc Chem Res 2020;53:1046-55. [Crossref] [PubMed]
- Narayanankutty A, Sasidharan A, Job JT. Targeting Toll like Receptors in Cancer: Role of TLR Natural and Synthetic Modulators. Curr Pharm Des 2020;26:5040-53. [Crossref] [PubMed]
- Damiano V, Rosa R, Formisano L, et al. Toll-like receptor 9 agonist IMO cooperates with everolimus in renal cell carcinoma by interfering with tumour growth and angiogenesis. Br J Cancer 2013;108:1616-23. [Crossref] [PubMed]
- Rini BI, Battle D, Figlin RA, et al. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of advanced renal cell carcinoma (RCC). J Immunother Cancer 2019;7:354. [Crossref] [PubMed]
- Schadendorf D, Hodi FS, Robert C, et al. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J Clin Oncol 2015;33:1889-94. [Crossref] [PubMed]
- Vitale I, Shema E, Loi S, et al. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat Med 2021;27:212-24. [Crossref] [PubMed]
- Haist M, Stege H, Grabbe S, et al. The Functional Crosstalk between Myeloid-Derived Suppressor Cells and Regulatory T Cells within the Immunosuppressive Tumor Microenvironment. Cancers (Basel) 2021;13:210. [Crossref] [PubMed]
- Shen H, Liu J, Chen S, et al. Prognostic Value of Tumor-Associated Macrophages in Clear Cell Renal Cell Carcinoma: A Systematic Review and Meta-Analysis. Front Oncol 2021;11:657318. [Crossref] [PubMed]
- Sanders C, Hamad ASM, Ng S, et al. CD103+ Tissue Resident T-Lymphocytes Accumulate in Lung Metastases and Are Correlated with Poor Prognosis in ccRCC. Cancers (Basel) 2022;14:1541. [Crossref] [PubMed]
- Li Y, Wang Z, Jiang W, et al. Tumor-infiltrating TNFRSF9+ CD8+ T cells define different subsets of clear cell renal cell carcinoma with prognosis and immunotherapeutic response. Oncoimmunology 2020;9:1838141. [Crossref] [PubMed]
- McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 2019;37:457-95. [Crossref] [PubMed]
- Zhang Z, Liu S, Zhang B, et al. T Cell Dysfunction and Exhaustion in Cancer. Front Cell Dev Biol 2020;8:17. [Crossref] [PubMed]
- Derynck R, Turley SJ, Akhurst RJ. TGFβ biology in cancer progression and immunotherapy. Nat Rev Clin Oncol 2021;18:9-34. [Crossref] [PubMed]
- Sawant DV, Yano H, Chikina M, et al. Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. Nat Immunol 2019;20:724-35. [Crossref] [PubMed]


