
Genomic analysis of the endosomal sorting required for transport complex III pathway genes as therapeutic and prognostic biomarkers for endometrial carcinoma
Introduction
Endometrial carcinoma (EC) is one of the most common gynecological cancers worldwide, with an estimated new cases and deaths of 65,950 and 12,550, respectively (1). EC has two subgroups, namely, types I and II. Type I is characterized by an upregulation of hormonal receptors. It consists of 60–70% of EC and histological grades 1 and 2. Type II is characterized by low levels of estrogen and progesterone receptor expression. Moreover, it is associated with high-grade EC and serous or clear cell carcinoma with poor prognosis (2). After surgical resection, chemotherapy and radiotherapy for EC, the 5-year disease-free survival (DFS) of patients diagnosed in the early stages, stages III or IV, and recurrent or metastatic disease decreased progressively to 74.2–90.8%, 57.3–66.2%, 20.1–25.5%, and 16%, respectively (3). Targeted therapies for tyrosine kinases and immunotherapy using immune-checkpoint inhibitors (4) are clinical strategies used in cancer treatment. These modalities aim to kill tumor cells and protect normal cells.
The endosomal sorting required for transport complex III (ESCRT-III) acts as a protective mechanism that delays cell death by repairing damaged plasma membranes due to necroptosis (5), pyroptosis (6), and ferroptosis (7). The ESCRT-III complex incorporates charged multivesicular body protein 2A (CHMP2A), CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP5, and CHMP7, which belong to the chromatin-modifying protein/charged multivesicular body protein (CHMP) family (8) (Figure S1). These are involved in the MVB sorting pathway and are important in recycling and degradation of membrane proteins. ESCRT-I, ESCRT-II, and ESCRT-III, as well as VPS4A and ALIX were triggered by Ca2+ influx, recruited to damaged lysosomes and mediated lysosomal membrane repair (8). Detailed information on the location and synonyms of these eight ESCRT-III genes is listed in Tables S1,S2. Since regulated cell death usually occurs in drug resistant tissues during EC therapy (9), we aimed to determine whether ESCRT-III genes might function as potential targets in the treatment and prognosis of EC.
Searching for tumor genomic biomarker using microarray RNA sequencing (RNA-Seq) data was recently applied to determine cancer prognosis. In this study, we performed bioinformatics technology with a high-throughput screening data to identify the ESCRT-III pathway gene signature in EC using the UALCAN, Clinical Proteomic Tumor Analysis Consortium (CPTAC) Confirmatory/Discovery, TISIDB, Gene Expression Profiling Interactive Analysis (GEPIA), and NIH National Cancer Institute CDC Data Portal dataset on the uterine corpus endometrial carcinoma (UCEC) dataset of the Cancer Genome Atlas (TCGA) database. Data on gene expression profiles, pathological relationship, somatic mutation status, function enrichment pathway, correlation with tumor infiltrating lymphocytes (TILs), and survival analysis were generated to assess if ESCRT-III pathway genes could be regarded as potential therapeutic targets and candidate prognostic biomarkers for EC. We present the following article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-660/rc).
Methods
Acquisition of data on ESCRT-III pathway genes
This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). Data on the transcriptional levels of over- and under-expressed ESCRT pathway genes as determined by Transcriptome RNA sequence (RNA-Seq) analysis were collected using the UALCAN analysis (http://ualcan.path.uab.edu) (EC: 546; normal: 35) (10) from the UCEC dataset of the TCGA database (https://portal.gdc.cancer.gov/). We also conducted GEPIA (http://gepia.cancer-pku.cn/) (11) for data mining and visualization of the RNA sequencing expression data to validate the expression profiles of the ESCRT pathway genes in UCEC primary tumors (n=174) from TCGA-UCEC and GTEx-Uterus projects (12). The matched TCGA normal and GTEx data were combined into the normal group (n=91). The default parameters were |Log2FC| Cutoff of 1, P value cutoff of 0.01, and log2 (transcripts per million, TPM +1) for log-scale were used. A jitter size of 0.4 was generated. Z-values representing standard deviations from the median across EC and normal endometrial tissue samples were identified using the Clinical Proteomic Tumor Analysis Consortium (CPTAC) Confirmatory/Discovery dataset (http://ualcan.path.uab.edu/analysis-prot.html) (13) (EC: 100; normal: 31) and normalized by Log2 spectral count ratio values both in each sample profile and across samples.
Clinical pathological relationship and pathway analyses of the ESCRT genes
The relationship between ESCRT genes and basic clinical pathological information, including subtype, tumor grade, histology, individual cancer stages, and methylation status, were further explored using the UALCAN and CPTAC datasets. We systematically assessed the ESCRT genes involved in key pathways across EC samples based on the CPTAC dataset, which were represented by Z-values. Associations of the ESCRT-III pathway gene expression and immune/molecular subtypes across EC analyzed using the Kruskal-Wallis Test -log10(P value) were identified on the TISIDB (http://cis.hku.hk/TISIDB/index.php) (14) dataset.
Function enrichment and pathway analyses of the ESCRT pathway genes
To obtain the enriched biological functions and pathways of the ESCRT pathway genes, we conducted the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses on TISIDB. Statistical significance was set at P<0.05. Connections among the ESCRT pathway gene expression were analyzed via the Pearson’s correlation on the GEPIA dataset.
Validation of ESCRT gene mutations in the EC samples
We obtained the mutation data on the ESCRT pathway genes in the TCGA-UCEC cohort from the NIH National Cancer Institute GDC Data Portal database (https://portal.gdc.cancer.gov). We calculated the number of cases of ESCRT genes affected by simple somatic mutation (SSM) across the TCGA project (n=530). Cases affected by copy number variation (CNV) gains and losses in the TCGA database (n=510) were also analyzed computationally. Disease type in the UCEC project included adenomas, adenocarcinomas, cystic, mucinous, serous, and epithelial neoplasms. Statistical calculations of mutation types, including somatic mutations, CNV losses, CNV gains, SSM affected, variant effect predictor (VEP) impact mutation, Sorting Intolerant From Tolerant (SIFT) impact mutation, Polyphen Impact Mutation, Consequence Type Mutation, Type Mutation and Variant Caller Mutation were obtained and listed in Table S3. The frequency and pathogenicity of the variants in the exons were determined using bioinformatic prediction tools. We also gathered the clinical survival time and status of the EC patients.
Association between gene expression and TILs
We utilized the online tools, TISIDB and GEPIA2021 (http://gepia2021.cancer-pku.cn/sub-expression.html), based on the TCGA database using Spearman correlations to investigate the correlations between genes and their immune microenvironment (15). The relative abundance of 28 TIL types was determined using gene set variation analysis (GSVA) based on gene expression profiles (16).
Survival analysis
To screen the relationship between the expression profiles of the ESCRT pathway genes and the overall survival (OS) between high- and low-expression groups from the TCGA-UCEC dataset, we inferred the Kaplan-Meier survival curves on UALCAN [N (high vs. low/medium) =137 vs. 408] and TISDB analysis. Genes with a statistical significance, as determine by conducting the log-rank test (P<0.05), were considered prognostic genes of EC. We also carried out a univariate Cox proportional hazards regression analysis in GEPIA with EC samples that had high and low expression levels in 86 cases from the TCGA to evaluate their OS and DFS. The Cox proportional hazard ratio (HR) was based on the Cox PH model, while the 95% confidence interval (CI) was included in the survival plot as a dotted line. The median expression threshold for splitting the high- and low-expression cohorts as group cutoff was established. Patients who exhibited an upregulation showed expression values > third quartile. They were then divided into high- or low-expression groups with a median group cutoff of 50%. ESCRT gene mutations associated with survival rates were collected.
Statistical analysis
RNA-seq expression of the TPM of ESCRT pathway genes in the TCGA-UCEC and GTEx-Uterus data, clinicopathological relationships, and pathways were analyzed via ANOVA. The ESCRT pathway genes associated with immune and molecular subtypes were analyzed using the Kruskal-Wallis test -log10(P value). The Chi-square and the Fisher’s exact tests were used to compare the mutations. The Spearman’s correlation test was used to explore the correlation between gene expression levels and the immune cell types. The Kaplan-Meier survival curves were used to analyze the OS, and univariate Cox proportional hazards regression analysis was used to evaluate the OS and DFS. The statistical significance was set at P<0.05.
Online database
The online databases were as follows: TCGA (https://portal.gdc.cancer.gov/); UALCAN (http://ualcan.path.uab.edu) (10); TISIDB (http://cis.hku.hk/TISIDB/index.php (14); GEPIA: Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/) (11); GEPIA2021 (http://gepia2021.cancer-pku.cn/sub-expression.html; CPTAC Confirmatory/Discovery dataset (http://ualcan.path.uab.edu/analysis-prot.html) (13); and NIH National cancer institute CDC Data Portal database (https://portal.gdc.cancer.gov).
Results
Validation of the ESCRT-III pathway genes in EC samples
We investigated the transcriptional RNA expression levels of the key genes in the ESCRT-III pathway in EC and normal endometrial tissues. CHMP2B, CHMP5, CHMP5, and CHMP7 were significantly lower, while CHMP2A and CHMP4C were significantly higher in EC samples (n=546) than in normal endometrial samples (n=35) in the UALCAN analysis of the TCGA-UCEC database (Figure S2A, Table S4). We also verified the expression values of the ESCRT genes by conducting the GEPIA analysis, which indicated a significantly higher expression level of CHMP4C in the EC samples (n=174) than in the normal samples (n=91) on the TCGA-UCEC and GTEx-Uterus data (n=13). Genes, such as CHMP2A, CHMP4B, and CHMP5, have higher levels of expression (Figure S2B, Table S4). On the other hand, CHMP2B, CHMP3, CHMP5 and CHMP7 had lower levels of expression in the EC samples. In addition, differential analysis through CPTAC characterized by Z-values revealed that CHMP2A, CHMP2B, CHMP3, and CHMP4B were markedly lower in the EC samples (n=100) and normal endometrial tissues (n=31), whereas CHMP4C and CHMP5 showed significantly higher differences in EC samples (Figure S2C, Table S4). CHMP2A had a higher level of expression on the UALCAN dataset than on the CPTAC. CHMP5 had a significantly lower expression on the UALCAN dataset than on the CPTAC. This may possibly be due to different sample sizes and comparison methods with transcriptional RNA expression levels on UALACN, GEPIA, and the Z-value of standard deviations on CPTAC.
Analysis of the clinical pathological characteristics of the ESCRT-III pathway genes in EC samples
We further compared the expression profiles of ESCRT pathway genes in different tumor stages, grades, subtypes, histological/immune/molecular subtypes, TP53 mutants, methylation, and pathways altered on the TCGA-UCEC and GTEx-Uterus datasets in the UALCAN, CPTAC, and TISDIB analyses, respectively. In the UALCAN analysis, CHMP2A, CHMP2B, CHMP4C, CHMP5, and CHMP7 were significantly differentially expressed among the International Federation of Gynecology and Obstetrics (FIGO) stages 1 to 3 and the normal sample. Different analyses revealed a significantly increased expression of CHMP2A, CHMP4C, and CHMP7, as well as decreased expression of CHMP2B and CHMP5 in endometrioid and serous carcinoma samples compared with normal samples. In addition, CHMP2A, CHMP4B, CHMP5, and CHMP7 were significantly different between the endometrioid and serous carcinoma samples. An increase in the expression of CHMP2A and CHMP7 and a decrease in the expression of CHMP4B were observed in the EC samples compared with the serous carcinoma samples. CHMP2A, CHMP4B, CHMP4C, CHMP5, and CHMP7 were significantly associated with TP53 mutation status in EC samples. Moreover, we found that the methylation status of CHMP2A, CHMP2B, CHMP4C, and CHMP5 was strongly correlated with their expression levels (Figure 1A, Table S4). In summary, we found that CHMP2B, CHMP3, CHMP4B, CHMP5, and CHMP7 were significantly downregulated, while CHMP2A, CHMP4C were significantly upregulated in the EC samples.
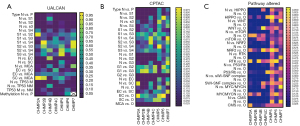
In the CPTAC analysis, all ESCRT pathway genes were significantly differentially expressed between histologic grades 2 and 3. CHMP2A, CHMP2B, CHMP3, CHMP4B, and CHMP4C were significantly differentially expressed between grade 1, grade 2, and normal samples. To investigate the correlations between the pathway alterations, CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, and CHMP5 were differentially expressed between endometrial cancer tissues (n=100) and normal endometrial tissues (n=31) in HIPPO, WNT, mTOR, NRF2, RTK, P53/Rb, SWI-SNF, MYC/MYCN pathway, and chromatin modifier status. This was determined by conducting the Pearson correlation analysis (Figure 1B, Table S4).
TISDIB analysis showed that CHMP2A, CHMP4B, and CHMP5 were significantly associated with tumor stage, while CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, and CHMP7 were significantly differentially expressed among different tumor grades. Except for CHMP7, all other ESCRT genes were significantly associated with immune subtypes, including wound healing, IFN-gamma dominant, inflammatory, lymphocyte depleted, immunologically quiet, and TGF-b dominant. This was determined by performing the Kruskal-Wallis test (Figure S3). CHMP2A, CHMP4B, CHMP5, and CHMP7 were significantly associated with a molecular subtype, including POLE (DNA polymerase epsilon), low copy number (CN-LOW), high copy number (CN-HIGH), and microsatellite instability (MSI) (Table S4).
Functional enrichment analysis
GO and KEGG enrichment analyses were performed to determine the biological significance of the genes involved in the ESCRT pathway. Endocytosis (I04144) was the most significantly enriched KEGG pathway analysis, which showed that ESCRT genes were significantly enriched in the endosomal sorting complex required for transport (ESCRT) (hsa917729), membrane trafficking (R-HAS-199991), vesicle-mediated transport (R-HAS-5653656), and macroautophagy (R-HAS-1632852). The main cellular components (CC) were the ESCRT complex (GO:0036452), ESCRT III complex (GO:0000815), endosome membrane (GO:0010008), late endosome (GO:0005770), late endosome membrane (GO:0031902), and endosomal part (GO:0044440).
The GO analysis demonstrated that all the ESCRT pathway genes (CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP5, and CHMP7) were mainly involved in the biological process (BP) of membrane budding (GO:0006900), endosome organization (GO:0007032), endosomal transport (GO:0016197), multivesicular body organization (GO:0036257), multivesicular body assembly (GO:0036258), multi-organism membrane organization (GO:0044803), and multi-organism membrane budding (GO:1902592), which were related to the main ESCRT function. In addition, CHMP5 participated in the ESCRT complex assembly (GO:1904895), ESCRT III complex assembly (GO:1904902), CHMP2A, CHMP5, and CHMP7, which participated in the ESCRT complex disassembly (GO:1904896) and ESCRT III complex disassembly (GO:1904903). In addition, CHMP3 was involved in the regulation of early endosome to late endosome transport (GO:2000641). On the other hand, CHMP2B, CHMP3, and CHMP5 were involved in the endosome-to-lysosome transport (GO:0008333). CHMP3 and CHMP5 were involved in the multivesicular body sorting pathway (GO:0071985). CHMP4B was involved in the protein localization to the membrane (GO:0072657), membrane fission (GO:0090148), and the establishment of protein localization to the membrane (GO:0090150) (Figure 1C). Detailed information on the above mentioned involvements is shown in Table 1.
Table 1
| Symbol | ||||||||
|---|---|---|---|---|---|---|---|---|
| CHMP2A | CHMP2B | CHMP3 | CHMP4B | CHMP4C | CHMP5 | CHMP6 | CHMP7 | |
| Project | ||||||||
| SSM (N=530) | ||||||||
| SSM affected cases | 21 | 26 | 15 | 22 | 14 | 19 | 9 | 36 |
| SSM affected cases in TCGA % | 0.0396 | 0.0491 | 0.0283 | 0.0415 | 0.0264 | 0.0358 | 0.017 | 0.0679 |
| CNV (N=510) | ||||||||
| CNV gains | 26 | 11 | 8 | 22 | 14 | 21 | 53 | 3 |
| CNV gains in TCGA % | 0.0510 | 0.0216 | 0.0157 | 0.0431 | 0.0275 | 0.0412 | 0.1039 | 0.0059 |
| CNV losses | 23 | 23 | 9 | 11 | 5 | 5 | 7 | 32 |
| CNV losses in TCGA % | 0.0451 | 0.0451 | 0.0176 | 0.0216 | 0.0098 | 0.0098 | 0.0137 | 0.0627 |
| All SSM affected cases in UCEC | 70 | 59 | 32 | 55 | 31 | 45 | 28 | 68 |
| Somatic mutation | 22 | 35 | 16 | 24 | 19 | 22 | 9 | 43 |
| Somatic mutation in all SSM affected cases in UCEC % | 0.3143 | 0.5932 | 0.5 | 0.4364 | 0.6129 | 0.4888 | 0.3214 | 0.6324 |
| Disease type | ||||||||
| Adenomas and adenocarcinomas | 32 | 31 | 18 | 32 | 21 | 26 | 28 | 40 |
| Cystic, mucinous and serous neoplasms | 38 | 28 | 13 | 23 | 10 | 19 | 0 | 28 |
| Epithelial neoplasms | 1 | |||||||
| Survival | ||||||||
| Dead | 13 | 10 | 5 | 10 | 8 | 14 | 16 | 9 |
| Alive | 57 | 49 | 27 | 45 | 23 | 31 | 51 | 59 |
| Survival rate | 0.82 | 0.72 | 0.81 | 0.76 | 0.68 | 0.65 | 0.25 | 0.84 |
| Interval of last follow-up (Year) | 5.095 | 5.142 | 5.136 | 5.106 | 5.106 | 5.095 | 5.008 | 5.106 |
ESCRT, endosomal sorting complex required for transport; TCGA, The Cancer Genome Atlas; CNV, copy-number variant; SSM, simple somatic mutation; SNV, single nucleotide variation; UCEC, uterine corpus endometrial carcinoma.
The Pearson correlation coefficient analysis results demonstrated that all of the concerned genes enriched in the ESCRIhsa917729) KEGG pathway in the GEPIA dataset were connected with each other significantly by transcripts per million (TPM) expression values (Figure S4).
ESCRT gene mutations in the EC samples
We observed seven ESCRT pathway genes that were most frequently mutated in the EC samples obtained from the simple somatic mutation (SSM) TCGA cohort (n=530), which included various cancer types, including CHMP2A (3.96%), CHMP2B (4.91%), CHMP3 (2.83%), CHMP4B (4.15%), CHMP4C (2.64%), CHMP5 (3.58%), and CHMP7 (6.97%). CHMP5 (1.70%) was ranked third on the GDC Data Portal (Figure S5). Different mutation categories of the ESCRT genes in the TCGA-UCEC project showed that moderate somatic mutations and missense mutations were the most common types (Figure 2A), CHMP7 ranked most of the various mutations type. Also, CHMP5 firstly showed CNV gains (Figure 2B). Moreover, the ESCRT pathway genes frequently mutated in the modifier of VEP impact mutation (Figure 2C), deleterious and tolerated of SIFT impact mutation (Figure 2D), probably_damaging and benign of polyphen impact mutation (Figure 2E), missense_variant, downstream_gene_variant, 3_prime_UTR_variant of consequence type mutation (Figure 2F), single base substitution of type mutation (Figure 2G) and somaticsniper mutation (Figure 2H) (Table 2, Table S3).
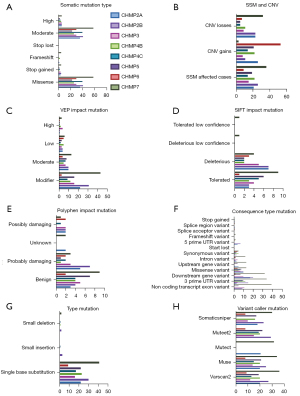
Table 2
| Ontology ID | Description | Genes |
|---|---|---|
| BP | ||
| GO:0000045 | Autophagosome assembly | CHMP4B |
| GO:0000070 | Mitotic sister chromatid segregation | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0000226 | Microtubule cytoskeleton organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0000281 | Mitotic cytokinesis | CHMP4B, |
| GO:0000819 | Sister chromatid segregation | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0000910 | Cytokinesis | CHMP4B, CHMP4C, |
| GO:0000920 | Cell separation after cytokinesis | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0001881 | Receptor recycling | CHMP5 |
| GO:0001894 | Tissue homeostasis | CHMP4B |
| GO:0001919 | Regulation of receptor recycling | CHMP5 |
| GO:0006612 | Protein targeting to membrane | CHMP4B |
| GO:0006620 | Posttranslational protein targeting to membrane | CHMP4B |
| GO:0006887 | Exocytosis | CHMP2A, |
| GO:0006900 | Membrane budding | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0006914 | Autophagy | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP6 |
| GO:0006997 | Nucleus organization | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0006998 | Nuclear envelope organization | CHMP2A, CHMP4B, CHMP7 |
| GO:0007032 | Endosome organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0007033 | Vacuole organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0007034 | Vacuolar transport | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0007040 | Lysosome organization | CHMP5 |
| GO:0007041 | Lysosomal transport | CHMP2B, CHMP5 |
| GO:0007051 | Spindle organization | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:0007059 | Chromosome segregation | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0007067 | Mitotic nuclear division | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0007080 | Mitotic metaphase plate congression | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0007088 | Regulation of mitotic nuclear division | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:0007098 | Centrosome cycle | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0007099 | Centriole replication | CHMP2A |
| GO:0007346 | Regulation of mitotic cell cycle | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:0008333 | Endosome to lysosome transport | CHMP2B, CHMP3, CHMP5 |
| GO:0009838 | Abscission | CHMP4C |
| GO:0010324 | Membrane invagination | CHMP2A |
| GO:0010458 | Exit from mitosis | CHMP2A, CHMP4B, CHMP7 |
| GO:0010506 | Regulation of autophagy | CHMP4B |
| GO:0010507 | Negative regulation of autophagy | CHMP4B |
| GO:0010639 | Negative regulation of organelle organization | CHMP2A, CHMP4B |
| GO:0010824 | Regulation of centrosome duplication | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0010826 | Negative regulation of centrosome duplication | CHMP2A |
| GO:0010948 | Negative regulation of cell cycle process | CHMP2A, CHMP4C |
| GO:0016050 | Vesicle organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0016197 | Endosomal transport | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0016236 | Macroautophagy | CHMP3, CHMP4B |
| GO:0016241 | Regulation of macroautophagy | CHMP4B |
| GO:0016242 | Negative regulation of macroautophagy | CHMP4B |
| GO:0016482 | Cytosolic transport | CHMP3 |
| GO:0017157 | Regulation of exocytosis | CHMP2A |
| GO:0019058 | Viral life cycle | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0019068 | Virion assembly | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0019076 | Viral release from host cell | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:0031023 | Microtubule organizing center organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0031468 | Nuclear envelope reassembly | CHMP4B, CHMP7 |
| GO:0032465 | Regulation of cytokinesis | CHMP4C |
| GO:0032466 | Negative regulation of cytokinesis | CHMP4C |
| GO:0032886 | Regulation of microtubule-based process | CHMP4B, CHMP5 |
| GO:0032386 | Regulation of intracellular transport | CHMP3 |
| GO:0032509 | Endosome transport via multivesicular body sorting pathway |
CHMP2B, CHMP3 |
| GO:0032510 | Endosome to lysosome transport via multivesicular body sorting pathway |
CHMP2B, CHMP3 |
| GO:0032886 | Regulation of microtubule-based process | CHMP2A, CHMP2B, CHMP3, CHMP4B |
| GO:0032984 | Macromolecular complex disassembly | CHMP2A, CHMP2B, CHMP5, CHMP7 |
| GO:0036257 | Multivesicular body organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0036258 | Multivesicular body assembly | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0039702 | Viral budding via host ESCRT complex | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP6, CHMP7 |
| GO:0043112 | Receptor metabolic process | CHMP5 |
| GO:0043241 | Protein complex disassembly | CHMP2A, CHMP2B, CHMP5, CHMP7 |
| GO:0043900 | Regulation of multi-organism process | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:0043901 | Negative regulation of multi-organism process | CHMP3 |
| GO:0043902 | Positive regulation of multi-organism process | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:0043903 | Regulation of symbiosis, encompassing mutualism through parasitism |
CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:0044770 | Cell cycle phase transition | CHMP2A, CHMP4B, CHMP7 |
| GO:0044772 | Mitotic cell cycle phase transition | CHMP2A, CHMP4B, CHMP7 |
| GO:0044801 | Single-organism membrane fusion | CHMP2B, CHMP3 |
| GO:0044803 | Multi-organism membrane organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0045022 | Early endosome to late endosome transport | CHMP3 |
| GO:0045047 | Protein targeting to ER | CHMP4B |
| GO:0045324 | Late endosome to vacuole transport | CHMP7 |
| GO:0045786 | Negative regulation of cell cycle | CHMP2A, CHMP4C |
| GO:0045921 | Positive regulation of exocytosis | CHMP2A |
| GO:0046599 | Regulation of centriole replication | CHMP2A |
| GO:0046600 | Negative regulation of centriole replication | CHMP2A |
| GO:0046605 | Regulation of centrosome cycle | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0046606 | Negative regulation of centrosome cycle | CHMP2A |
| GO:0046755 | Viral budding | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0048284 | Organelle fusion | CHMP2B, CHMP3, CHMP4C |
| GO:0048524 | Positive regulation of viral process | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:0048871 | Multicellular organismal homeostasis | CHMP4B |
| GO:0050000 | Chromosome localization | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0048525 | Negative regulation of viral process | CHMP3 |
| GO:0050792 | Regulation of viral process | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:0050890 | Cognition | CHMP2B |
| GO:0051047 | Positive regulation of secretion | CHMP2A |
| GO:0051225 | Spindle assembly | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:0051258 | Protein polymerization | CHMP2A, CHMP3 |
| GO:0051259 | Protein oligomerization | CHMP2A, CHMP3, CHMP4B |
| GO:0051260 | Protein homooligomerization | CHMP2A, CHMP4B |
| GO:0051291 | Protein heterooligomerization | CHMP2A, CHMP3 |
| GO:0051297 | Centrosome organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0051298 | Centrosome duplication | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0051302 | Regulation of cell division | CHMP4C |
| GO:0051303 | Establishment of chromosome localization | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0051310 | Metaphase plate congression | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0051493 | Regulation of cytoskeleton organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0051494 | Negative regulation of cytoskeleton organization | CHMP2A |
| GO:0060627 | Regulation of vesicle-mediated transport | CHMP3 |
| GO:0051640 | Organelle localization | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0051656 | Establishment of organelle localization | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0051782 | Negative regulation of cell division | CHMP4C |
| GO:0051783 | Regulation of nuclear division | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:0060249 | Anatomical structure homeostasis | CHMP2B, CHMP4B |
| GO:0060627 | Regulation of vesicle-mediated transport | CHMP2A |
| GO:0061025 | Membrane fusion | CHMP2B, CHMP3 |
| GO:0061511 | Centriole elongation | CHMP2A |
| GO:0061763 | Multivesicular body-lysosome fusion | CHMP2B, CHMP3 |
| GO:0070050 | Neuron cellular homeostasis | CHMP2B |
| GO:0070507 | Regulation of microtubule cytoskeleton organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| GO:0070972 | Protein localization to endoplasmic reticulum | CHMP4B |
| GO:0070997 | Neuron death | CHMP4B |
| GO:0071985 | Multivesicular body sorting pathway | CHMP3, CHMP5 |
| GO:0072599 | Establishment of protein localization to endoplasmic reticulum | CHMP4B |
| GO:0072657 | Protein localization to membrane | CHMP4B |
| GO:0080171 | Lytic vacuole organization | CHMP5 |
| GO:0090148 | Membrane fission | CHMP4B |
| GO:0090150 | Establishment of protein localization to membrane | CHMP4B |
| GO:0090169 | Regulation of spindle assembly | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:0090174 | Organelle membrane fusion | CHMP2B, CHMP3 |
| GO:0090224 | Regulation of spindle organization | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:0090307 | Mitotic spindle assembly | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:0090611 | Ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway | CHMP4B, CHMP4C |
| GO:0097352 | Autophagosome maturation | CHMP3 |
| GO:0097576 | Vacuole fusion | CHMP2B, CHMP3 |
| GO:0098534 | Centriole assembly | CHMP2A |
| GO:0098813 | Nuclear chromosome segregation | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0098927 | Vesicle-mediated transport between endosomal compartments | CHMP3 |
| GO:1901673 | Regulation of mitotic spindle assembly | CHMP2A, CHMP2B, CHMP4C, CHMP5 |
| GO:1901214 | Regulation of neuron death | CHMP4B |
| GO:1902115 | Regulation of organelle assembly | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:1902116 | Negative regulation of organelle assembly | CHMP2A, CHMP4B |
| GO:1902186 | Regulation of viral release from host cell | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:1902187 | Negative regulation of viral release from host cell | CHMP3 |
| GO:1902188 | Positive regulation of viral release from host cell | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:1902590 | Multi-organism organelle organization | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:1902592 | Multi-organism membrane budding | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:1903335 | Regulation of vacuolar transport | CHMP3 |
| GO:1903649 | Regulation of cytoplasmic transport | CHMP3 |
| GO:1902850 | Microtubule cytoskeleton organization involved in mitosis | CHMP2A, CHMP2B, CHMP4B, CHMP4C, CHMP5 |
| GO:1903532 | Positive regulation of secretion by cell | CHMP2A |
| GO:1903541 | Regulation of exosomal secretion | CHMP2A |
| GO:1903543 | Positive regulation of exosomal secretion | CHMP2A |
| GO:1903722 | Regulation of centriole elongation | CHMP2A |
| GO:1903723 | Negative regulation of centriole elongation | CHMP2A |
| GO:1903900 | Regulation of viral life cycle | CHMP2A, CHMP2B, CHMP3, CHMP4C |
| GO:1903901 | Negative regulation of viral life cycle | CHMP3 |
| GO:1903902 | Positive regulation of viral life cycle | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C |
| GO:1904895 | ESCRT complex assembly | CHMP6 |
| GO:1904896 | ESCRT complex disassembly | CHMP2A, CHMP5, CHMP7 |
| GO:1904902 | ESCRT III complex assembly | CHMP6 |
| GO:1904903 | ESCRT III complex disassembly | CHMP2A, CHMP5, CHMP7 |
| GO:1905037 | Autophagosome organization | CHMP4B |
| GO:1990182 | Exosomal secretion | CHMP2A |
| GO:2000641 | Regulation of early endosome to late endosome transport | CHMP3 |
| GO:2000785 | Regulation of autophagosome assembly | CHMP4B |
| MF | ||
| GO:0002020 | Protease binding | CHMP3 |
| GO:0031210 | Phosphatidylcholine binding | CHMP2A, CHMP3 |
| GO:0045296 | Cadherin binding | CHMP2B, CHMP4B, CHMP5 |
| GO:0047485 | Protein N-terminus binding | CHMP6 |
| GO:0050839 | Cell adhesion molecule binding | CHMP2B, CHMP4B, CHMP5 |
| GO:0008565 | Protein transporter activity | CHMP7 |
| GO:0098631 | Protein binding involved in cell adhesion | CHMP2B, CHMP4B, CHMP5 |
| GO:0098632 | Protein binding involved in cell-cell adhesion | CHMP2B, CHMP4B, CHMP5 |
| GO:0098641 | Cadherin binding involved in cell-cell adhesion | CHMP2B, CHMP4B, CHMP5 |
| GO:0070405 | Ammonium ion binding | CHMP2A, CHMP3 |
| GO:1990381 | Ubiquitin-specific protease binding | CHMP3 |
| CC | ||
| GO:000815 | ESCRT III complex | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP6, CHMP7 |
| GO:0005635 | Nuclear envelope | CHMP2A, CHMP4B, CHMP7 |
| GO:0005770 | Late endosome | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP6 |
| GO:0005913 | Cell-cell adherens junction | CHMP5 |
| GO:0010008 | Endosome membrane | CHMP2A, CHMP2B, CHMP3, CHMP4C, CHMP5, CHMP6, CHMP7 |
| GO:0030117 | Membrane coat | CHMP2A |
| GO:0030496 | Midbody | CHMP4C |
| GO:0031902 | Late endosome membrane | CHMP2A, CHMP2B, CHMP3, CHMP4C, CHMP6 |
| GO:0036452 | ESCRT complex | CHMP2A, CHMP2B, CHMP3, CHMP4C, CHMP6, CHMP7 |
| GO:0044440 | Endosomal part | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP7 |
| GO:0048475 | Coated membrane | CHMP2A, CHMP4B |
| GO:0090543 | Flemming body | CHMP4C |
| GO:0098552 | Side of membrane | CHMP4B |
| GO:0098562 | Cytoplasmic side of membrane | CHMP4B |
| KEGG pathway | ||
| Hsa04144 | Endocytosis | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5 |
| Reactome pathway | ||
| R-HAS-162588 | Budding and maturation of HIV virion | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-2262752 | Cellular responses to stress | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP6, CHMP7 |
| R-HAS-421837 | Clathrin derived vesicle budding | CHMP2A |
| R-HAS-1643685 | Disease | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-917729 | Endosomal Sorting Complex Required For Transport (ESCRT) | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-162906 | HIV Infection | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-162587 | HIV Life Cycle | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-5663205 | Infectious disease | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-162599 | Late Phase of HIV Life Cycle | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-432720 | Lysosome Vesicle Biogenesis | CHMP2A |
| R-HAS-1632852 | Macroautophagy | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP6, CHMP7 |
| R-HAS-199991 | Membrane Trafficking | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-5653656 | Vesicle-mediated transport | CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, CHMP6, CHMP7 |
| R-HAS-199992 | Trans-Golgi Network Vesicle Budding | CHMP2A |
BP, biological process; CC, cellular component; ESCRT, MF, molecular function; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes; ER,endoplasmic reticulum; HIV, human immunodeficiency virus.
ESCRT genes associated with TILs
We explored the correlation between the expression of ESCRT genes and TILs, including 28 distinct immune cell types in the EC samples (n=546) in the TCGA-UCEC database with Spearman correlations using the online tools, TISIDB and GEPIA2021. We found that CHMP2A, CHMP3, CHMP4B, CHMP5, CHMP5, and CHMP7 were positively correlated with the infiltration of CD8. CHMP2B, CHMP3, CHMP4B, CHMP4C, CHMP5, and CHMP7 were positively correlated with the infiltration of CD4. CHMP4B, and CHMP5 were positively correlated with the infiltration of Tregs. CHMP3, CHMP4B, and CHMP5 were positively correlated with the infiltration of NK cells. On the other hand, CHMP2A and CHMP5 were negatively correlated with the infiltration of CD4. CHMP4C was negatively correlated with the infiltration of NK and Treg. CHMP2B and CHMP4C were negatively correlated with the infiltration of CD8, B cells, and macrophages. CHMP2A, CHMP3, CHMP5, and CHMP7 were negatively correlated with B cells (Figure 3A,3B, Table S5). We also detected CHMP2A (P=0.0081), CHMP4B (P=0.0005), CHMP4C (P=0.0002), CHMP5 (P=0.0231) and CHMP5 (P=0.0061) were significantly related with TILs (rho value) by multiple liner regression (Table 3). Significant differences between the ESCRT genes and TILs were observed in the TCGA-UCEC tumor, normal, and GTEx-uterus datasets in the GEPIA analysis, in which the parameters were grouped by tissue (P<0.05) (Figure 3C, Table S5). These results indicate that the ESCRT-III pathway genes may be effective markers in EC immunotherapy.
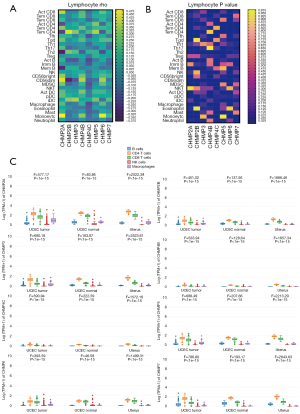
Table 3
| Model | CHMP2A | CHMP2B | CHMP3 | CHMP4B | CHMP4C | CHMP5 | CHMP6 | CHMP7 |
|---|---|---|---|---|---|---|---|---|
| Parameter estimates | 0.1175 | −0.002861 | 0.01904 | 0.1020 | −0.09654 | 0.05768 | 0.09136 | 0.01939 |
| Standard error | 0.04108 | 0.03143 | 0.02620 | 0.02584 | 0.02275 | 0.02394 | 0.03072 | 0.02259 |
| 95% CI | 0.03321 to 0.2018 | −0.06735 to 0.06163 | −0.03473 to 0.07280 | 0.04903 to 0.1550 | −0.1432 to −0.04985 | 0.008554 to 0.1068 | 0.02832 to 0.1544 | −0.02697 to 0.06575 |
| [t] | 2.860 | 0.09102 | 0.7264 | 3.950 | 4.243 | 2.409 | 2.974 | 0.8583 |
| P value | 0.0081 | 0.9281 | 0.4738 | 0.0005 | 0.0002 | 0.0231 | 0.0061 | 0.3983 |
TILs, tumor-infiltrating immune cells; ESCRT, endosomal sorting complex required for transport; CI, confidence interval.
Survival analysis and efficacy evaluation of the ESCRT genes
We performed the Kaplan-Meier survival analysis in UALCAN (Figure S6A). The patients were assigned to high (n=136) or low/medium (n=407) expression groups. The univariate Cox regression analysis in TISIDB (Figure S6B) from the TCGA-UCEC database was used to predict the prognostic values of the ESCRT genes on the OS of patients with EC. The Univariate Cox proportional hazards regression analysis in the GEPIA from TCGA also evaluated OS and DFS in patients with EC. We found that a low expression of CHMP4B (log-rank P=0.037) in UALCAN and a high expression of CHMP2A in TISDIB (log-rank P=0.0203) had a favorable OS for EC, while high expression of CHMP2A [(log-rank) P=0.016, hazard ratio (HR) (high) =0.44, P(HR) =]0.018) and CHMP7 [(log-rank) P=0.027, HR (high) =0.47, P(HR) =0.03] in GEPIA were positively correlated with a better prognosis of EC (Figure S6C).
In addition, we also performed a survival analysis using the Kaplan-Meier method on the ESCRT mutation genes; the 5-year OS rates/intervals of the last follow-up were as follows: CHMP2A (82%, 5.095), CHMP2B (72%, 5.142), CHMP3 (81%, 5.136), CHMP4B (76%, 5.106), CHMP4C (68%, 5.106), CHMP5 (65%, 5.095), CHMP5 (25%, 5.008), and CHMP7 (84%, 5.106) (Figure S7). The CHMP5 mutations positively correlated with a spoor prognosis. CHMP2A, CHMP3, and CHMP7 mutations demonstrated a good potential for being a prognostic tool in patients with EC (Figure S6, Table S6).
Discussion
Type I EC is associated with microsatellite instability hypermutated (MSI-H) and a copy-number low (CNL) integrated genomic categories in TCGA and frequent mutations in the mTOR pathway (2). Type II EC is characterized by a copy-number high (CN-H) and frequent mutation of TP53 (17). We found that CHMP2B, CHMP3, CHMP4B, CHMP5, CHMP5, and CHMP7 were significantly lower, whereas CHMP2A and CHMP4C were significantly higher in EC samples than in normal tissues. Since histological grade and clinical stage greatly affected the prognosis prediction of EC (1), we explored it found that all of the ESCRT pathway genes were significantly differentially expressed between grades 2 and 3. Increased expression of CHMP2A and CHMP7 and decreased expression of CHMP4B were related to endometrioid carcinoma samples compared with serous carcinoma samples. Furthermore, the results of the survival analysis revealed that upregulation of CHMP2A and CHMP7 and downregulation of CHMP4B corresponded to a good prognosis in patients with EC. All three genes were significantly associated with all four molecular subtypes in TCGA. Therefore, we indicated that high levels of CHMP2A and CHMP7 and low levels of CHMP4B were associated with good prognostic tumor type and early grade and were effective indicators of good prognosis in EC. CHMP2A, CHMP2B, CHMP3, CHMP4B, CHMP4C, and CHMP5 were significantly correlated with HIPPO, WNT, mTOR, NRF2, RTK, P53/Rb, SWI-SNF, MYC/MYCN pathway, and chromatin modifier status in EC tissues. As such, temsirolimus and everolimus, which are mTOR inhibitors, are effective in the treatment of recurrent chemotherapy (18,19). Moreover, ESCRT-III genes are effective markers for targeted therapy.
We found that most of the ESCRT pathway genes were frequently mutated in EC samples compared with the other cancer types obtained from the TCGA cohort. Additionally, missense mutations were the most common type of mutation. CHMP7 was the first modifier of VEP impact mutation, which was deleterious and tolerated by the SIFT Impact Mutation. Mutations in CHMP2A and CHMP7 had a good prognostic potential in EC. Dysfunction of ESCRT was associated with cancer, myopathy, and neurodegeneration. Missense mutations in CHMP4B were associated with posterior subcapsular or polar cataracts (20-22). Other studies showed that CHMP2B mutations that were linked to defects in the process of dissociation from ESCRT disrupted endosome-to-lysosome trafficking, degradation, and recycling. Thus, this resulted in defective mitochondria, imbalanced iron homeostasis, reactive oxygen species, neuronal damage (23,24), endocytic and autophagic defects (25-27) in autosomal dominant presenile dementia (28), amyotrophic lateral sclerosis, cortical basal degeneration (CBD) or familial frontotemporal lobar degeneration (FTLD) and frontotemporal dementia (FTD) (29).
The results of functional analysis revealed that the endocytosis and ESCRT was the most significantly enriched KEGG and Reactome pathways associated with the ESCRT genes in ECs. All concerned ESCRT pathway genes were mainly involved in the BPs of membrane budding, multivesicular body organization, and assembly. The main related CC were the ESCRT and ESCRT III complex. The latter was involved in transmembrane protein delivery and in the promotion of nuclear envelope sealing and mitotic spindle disassembly during the late endosome anaphase (30). Protein sorting from the endosome to the vacuole/lysosome transportation and degradation in eukaryotic cells required the formation of MVBs, which, in turn, required the sequential functioning of ESCRT, -I, -II, and -III complexes in four steps: cargo recognition and sorting, which was initiated by the binding of ESCRT-0 to the Phosphatidyl inositol 3-phosphate (PI3P) on endosomes; cargo sequestration, in which ESCRT-0 recruited ESCRT-I and interacted with ESCRT-II and -III; MVB vesicle formation, in which ESCRT-III proteins dissociated and assembled; and the ESCRT disassembly, in which the final step of the MVB pathway was formed (31-33). CHMP5 served as an acceptor for the ESCRT-II complex on the endosomal membranes in the ESCRT-III complex, thus participating in ESCRT and ESCRT III complex assembly in BP, while CHMP2A, CHMP5, and CHMP7 participated in the BP of ESCRT and ESCRT III complex disassembly. In the late stages of cytokinesis, CHMP3 was responsible for endosomal sorting/trafficking of the EGF receptor. CHMP4C was a key component of the cytokinesis checkpoint that delayed abscission and prevented both premature chromosome bridges of the cells and the accumulation of DNA damages (34,35). CHMP2B, CHMP3, and CHMP5 were involved in the BP of the multivesicular body sorting pathways and the regulation of early to late endosome transport. All of the key genes were significantly and positively correlated with each other, revealing that the ESCRT pathway might also be involved in the mechanism of the MVB pathway in EC.
On the other hand, tumor-infiltrating immune cells (TICs) modulated cancer cell functions in the tumor microenvironment (36). This could be a basis for immunotherapy, in which the endogenous immune response against tumor cells would be stimulated. Furthermore, this could be associated with the prognosis of patients with EC. ECs stimulate immune checkpoints and activate negative feedback mechanisms in a locally immunosuppressed environment to evade the immune system (37). Memory CD4+ T cells, activated NK cells, and dendritic cells all decreased with an increase in EC grade and stage, which was related to the occurrence of EC. However, high levels of Tregs indicate a good prognosis (38). Our preliminary results show strong correlations between all ESCRT genes and infiltration levels of multiple immune cell types in EC samples. CHMP2A and CHMP7 were positively correlated with the infiltration of CD4 and negatively correlated with B cells, while CHMP4B was associated with the infiltration of CD4, CD8, Treg, and NK cells. These three genes may potentially be immune-related indicators for the prognosis of EC and may become the basis for EC immunotherapies.
It has been found that EC cells represent the highest activated programmed death receptor-1 (PD1)/PD-L1expression in endometrioid endometrial adenocarcinoma (EEC), serous endometrial adenocarcinoma (ESC), and clear cell subtypes for 40–80%, 10–68%, and 23–69%, respectively. On the other hand, inactivate tumor-infiltrating CD4 and CD8 T cells are found in the tumor microenvironment (39). Targeting the PD1 pathway may be a great strategy in enhancing the antitumor immune response for EC treatment. Anti-PD-L1 antibodies, such as pembrolizumab, nivolumab, and atezolizumab, have been proven clinically effective in tumors with mismatch-repair deficiency in EC patients in clinical trials, with a 13–48% ORR, 19.0% PFS, and 68.8% OS rates (40-44). Pembrolizumab is an immune checkpoint blockade anti-PD1 antibody that has been used in patients with POLE-mutated (43) and MMR-deficient endometrial cancer (45). We observed that, except for CHMP7, all other ESCRT genes were significantly associated with the immune subtype. CHMP2A, CHMP4B, CHMP5, and CHMP7 were significantly associated with the POLE-mutated molecular subtype in TCGA, which was identified in 10% of endometrioid subtypes. Therefore, elucidation of CHMP2A, CHMP4B, CHMP7, and immune cell interplay would assist in the prediction of immunotherapy responses and development of novel immunotherapy targets.
Conclusions
Our present study confirmed that RNA expression data on multiple datasets showed the relationship between ESCRT-III genes and EC. ESCRT-III genes may be used as clinical biomarkers for the prognostic prediction and immunotherapy strategies in patients with EC. Further research should be conducted to determine the mechanisms of ESCRT genes as biomarkers and therapeutic targets for the treatment of EC.
Acknowledgments
We thank Elsevier Author Servives (https://webshop.elsevier.com/language-editing-services/language-editing/) for its linguistic assistance during the preparation of this manuscript.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-660/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-660/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-660/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol 1983;15:10-7. [Crossref] [PubMed]
- Connor EV, Rose PG. Management Strategies for Recurrent Endometrial Cancer. Expert Rev Anticancer Ther 2018;18:873-85. [Crossref] [PubMed]
- Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350-5. [Crossref] [PubMed]
- Gong YN, Guy C, Olauson H, et al. ESCRT-III Acts Downstream of MLKL to Regulate Necroptotic Cell Death and Its Consequences. Cell 2017;169:286-300.e16. [Crossref] [PubMed]
- Rühl S, Shkarina K, Demarco B, et al. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science 2018;362:956-60. [Crossref] [PubMed]
- Dai E, Meng L, Kang R, et al. ESCRT-III-dependent membrane repair blocks ferroptosis. Biochem Biophys Res Commun 2020;522:415-21. [Crossref] [PubMed]
- McCullough J, Frost A, Sundquist WI. Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes. Annu Rev Cell Dev Biol 2018;34:85-109. [Crossref] [PubMed]
- Liu J, Kang R, Tang D. ESCRT-III-mediated membrane repair in cell death and tumor resistance. Cancer Gene Ther 2021;28:1-4. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Zhong X, Liu Y, Liu H, et al. Identification of Potential Prognostic Genes for Neuroblastoma. Front Genet 2018;9:589. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Chen F, Chandrashekar DS, Varambally S, et al. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun 2019;10:5679. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73. [Crossref] [PubMed]
- Oza AM, Elit L, Tsao MS, et al. Phase II study of temsirolimus in women with recurrent or metastatic endometrial cancer: a trial of the NCIC Clinical Trials Group. J Clin Oncol 2011;29:3278-85. [Crossref] [PubMed]
- Slomovitz BM, Lu KH, Johnston T, et al. A phase 2 study of the oral mammalian target of rapamycin inhibitor, everolimus, in patients with recurrent endometrial carcinoma. Cancer 2010;116:5415-9. [Crossref] [PubMed]
- Zhang XH, Da Wang J, Jia HY, et al. Mutation profiles of congenital cataract genes in 21 northern Chinese families. Mol Vis 2018;24:471-7. [PubMed]
- Shiels A, Bennett TM, Knopf HL, et al. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet 2007;81:596-606. [Crossref] [PubMed]
- Wang X, Wang D, Wang Q, et al. Broadening the Mutation Spectrum in GJA8 and CHMP4B: Novel Missense Variants and the Associated Phenotypes in Six Chinese Han Congenital Cataracts Families. Front Med (Lausanne) 2021;8:713284. [Crossref] [PubMed]
- Zhang Y, Schmid B, Nikolaisen NK, et al. Patient iPSC-Derived Neurons for Disease Modeling of Frontotemporal Dementia with Mutation in CHMP2B. Stem Cell Reports 2017;8:648-58. [Crossref] [PubMed]
- Han JH, Ryu HH, Jun MH, et al. The functional analysis of the CHMP2B missense mutation associated with neurodegenerative diseases in the endo-lysosomal pathway. Biochem Biophys Res Commun 2012;421:544-9. [Crossref] [PubMed]
- Filimonenko M, Stuffers S, Raiborg C, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 2007;179:485-500. [Crossref] [PubMed]
- Isaacs AM, Johannsen P, Holm I, et al. Frontotemporal dementia caused by CHMP2B mutations. Curr Alzheimer Res 2011;8:246-51. [Crossref] [PubMed]
- Lu Y, Zhang Z, Sun D, et al. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol Cell 2013;52:264-71. [Crossref] [PubMed]
- Skibinski G, Parkinson NJ, Brown JM, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet 2005;37:806-8. [Crossref] [PubMed]
- Toft A, Roos P, Jääskeläinen O, et al. Serum Neurofilament Light in Patients with Frontotemporal Dementia Caused by CHMP2B Mutation. Dement Geriatr Cogn Disord 2020;49:533-8. [Crossref] [PubMed]
- Schöneberg J, Lee IH, Iwasa JH, et al. Reverse-topology membrane scission by the ESCRT proteins. Nat Rev Mol Cell Biol 2017;18:5-17. [Crossref] [PubMed]
- Mierzwa BE, Chiaruttini N, Redondo-Morata L, et al. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol 2017;19:787-98. [Crossref] [PubMed]
- Schöneberg J, Pavlin MR, Yan S, et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 2018;362:1423-8. [Crossref] [PubMed]
- Maity S, Caillat C, Miguet N, et al. VPS4 triggers constriction and cleavage of ESCRT-III helical filaments. Sci Adv 2019;5:eaau7198. [Crossref] [PubMed]
- Liu B, Guo S, Li GH, et al. CHMP4C regulates lung squamous carcinogenesis and progression through cell cycle pathway. J Thorac Dis 2021;13:4762-74. [Crossref] [PubMed]
- Neggers JE, Paolella BR, Asfaw A, et al. Synthetic Lethal Interaction between the ESCRT Paralog Enzymes VPS4A and VPS4B in Cancers Harboring Loss of Chromosome 18q or 16q. Cell Rep 2020;33:108493. [Crossref] [PubMed]
- Costa AC, Santos JMO, Gil da Costa RM, et al. Impact of immune cells on the hallmarks of cancer: A literature review. Crit Rev Oncol Hematol 2021;168:103541. [Crossref] [PubMed]
- Di Tucci C, Capone C, Galati G, et al. Immunotherapy in endometrial cancer: new scenarios on the horizon. J Gynecol Oncol 2019;30:e46. [Crossref] [PubMed]
- Chen B, Wang D, Li J, et al. Screening and Identification of Prognostic Tumor-Infiltrating Immune Cells and Genes of Endometrioid Endometrial Adenocarcinoma: Based on The Cancer Genome Atlas Database and Bioinformatics. Front Oncol 2020;10:554214. [Crossref] [PubMed]
- Herzog TJ, Arguello D, Reddy SK, et al. PD-1, PD-L1 expression in 1599 gynecological cancers: Implications for immunotherapy. Gynecol Oncol. 2015;137:204-5. [Crossref]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Ott PA, Bang YJ, Berton-Rigaud D, et al. Safety and Antitumor Activity of Pembrolizumab in Advanced Programmed Death Ligand 1-Positive Endometrial Cancer: Results From the KEYNOTE-028 Study. J Clin Oncol 2017;35:2535-41. [Crossref] [PubMed]
- Makker V, Rasco DW, Dutcus CE, et al. A phase Ib/II trial of lenvatinib (LEN) plus pembrolizumab (Pembro) in patients (Pts) with endometrial carcinoma. J Clin Oncol 2017;35:5598. [Crossref]
- Mehnert JM, Panda A, Zhong H, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest 2016;126:2334-40. [Crossref] [PubMed]
- Santin AD, Bellone S, Buza N, et al. Regression of Chemotherapy-Resistant Polymerase ε (POLE) Ultra-Mutated and MSH6 Hyper-Mutated Endometrial Tumors with Nivolumab. Clin Cancer Res 2016;22:5682-7. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]


