
Prognostic impact of mitofusin 2 expression in colon cancer
Introduction
Colorectal cancer (CRC), one of the leading causes of cancer-related deaths worldwide, has mortality rates in Europe, the USA, and worldwide of 45%, 35%, and 47.8%, respectively (1,2). Recently, following progress in chemotherapy, targeted therapy, and immunotherapy, the 5-year survival rate of patients with advanced CRC has improved. However, the increase in survival time afforded by comprehensive therapies is modest, and the 5-year survival rate for advanced CRC remains low (3). CRC prognosis is predominantly based on the tumor, node, and metastasis (TNM) stage, tumor location, and histologic subtypes. Few molecular biomarkers can predict patient prognosis and serve as therapeutic targets (4). Thus, it is essential to identify novel target biomarkers for CRC, especially advanced CRC.
Mitochondrial dysfunction and the activation of glycolysis are widely accepted as the hallmarks of cancer (5). Mitofusin 2 (MFN2) encodes a GTPase protein located on the mitochondrial outer membrane and is also known as the hyperplasia suppressor gene. It was initially identified in the vascular smooth muscle cells of spontaneously hypertensive rats (6). Previous studies showed that MFN2 disorders are involved in multiple conditions such as hypertension, Charcot-Marie-Tooth disease, obesity, diabetes, atherosclerosis, and cancer (7,8). MFN2 acts as a tumor suppressor in diverse cancers, including cervical cancer, hepatocellular carcinoma, pancreatic cancer, breast cancer, gastric cancer, and bladder cancer (9-11). However, the potential relationship between MFN2 and colon cancer has not been fully investigated to the best of our knowledge.
Previously, we showed that MFN2 is a hyperplasia suppressor gene in CRC cells, and MFN2 overexpression, mediated by an adenoviral vector, exerts an anti-proliferative effect by inducing G2/M-phase arrest in CRC cells (12). This study investigated the relationship between MFN2 expression and prognosis in human colon cancer using data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). We used gene set enrichment analysis (GSEA) to investigate the biological pathways in colon cancer linked to the MFN2 regulatory network. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-589/rc).
Methods
Data collection
Transcriptomic expression data of colon cancer and corresponding patient clinical information were downloaded from TCGA (https://portal.gdc.cancer.gov). We examined 470 instances of colon cancer and 41 normal samples to investigate the variation in MFN2 expression. We used samples with comprehensive clinical features in logistic regression and Cox analyses, and boxplots were drawn to visualize the correlation between MFN2 expression and the clinical stage of colon cancer. We performed Kaplan-Meier survival analysis using tumor data from TCGA colon cancer samples. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Immunohistochemistry (IHC) staining and single cell RNA-sequencing (RNA-seq) data processing
Immunohistochemical pictures of MFN2 protein expression studies were done in normal and CRC tissues from the HPA (http://www.proteinatlas.org/) to determine differences in MFN2 expression at the protein level. For IHC, the anti-body HPA030554 was employed. GEO datasets (https://www.ncbi.nlm.nih.gov/gds) were used to download single cell RNA-seq data associated with colon cancer (GSE139555 and GSE146771). To identify key cell types, researchers used the Seurat tool in R v.4.1.2 software. Principal component analysis (PCA) was performed using highly variable genes, and primary components were computed using t-distributed stochastic neighbor embedding (t-SNE) analysis.
GSEA
The GSEA program is a robust analytical tool used to evaluate gene expression data that is available as a free software package including an initial database of >1,000 biologically defined gene sets (13). We used GSEA software (version 4.1.0) to identify the biological roles and pathways that differed between sample groups with high and low MFN2 expression. Gene set permutations were conducted 1,000 times for each family-based analysis, with MFN2 expression as a phenotypic marker. Finally, we ranked all enriched pathways by the normalized enrichment score (NES), nominal P value, and false discovery rate (q-value).
Statistical analysis
We used R v.4.1.2 for all statistical analyses. A Wilcoxon signed-rank test was used to compare the MFN2 expression between cancerous and paracancerous tissues. Logistic regression analysis was employed to correlate the MFN2 status with clinicopathological parameters. We performed Kaplan-Meier analysis to ascertain survival and calculate the best gene cut-off values using the survminer package in R. Univariate Cox regression analysis was used to examine the relationships between overall survival (OS) and clinical variables. Finally, we performed multivariate Cox regression analyses to screen out the independent prognostic factors for colon cancer patients. Differences with a P value <0.05 were deemed statistically significant.
Results
Patient characteristics
We searched TCGA on 20 November 2021 for RNA-seq expression data and patient clinical information related to 511 colon cancer samples, including 470 cancerous and 41 paracancerous samples. Invalid data were removed from the final dataset. The study included data from 205 men and 174 women with colon cancer who ranged in age from 31 to 90 years (average age 66.5 years; median age 68 years). According to the American Joint Committee on Cancer (AJCC) TNM staging system, 66 (17.4%) cases were classified as stage I, 152 (40.1%) as stage II, 101 (26.6%) as stage III, and 60 (15.8%) as stage IV. The 9 (2.4%) cases were classified as stage T1, 66 (17.4%) as T2, 262 (69.1%) as T3, and 42 (11.1%) as T4. Among these colon cancer patients, lymph node metastasis (N1-N2) was found in 152 cases, and distant metastasis (M1) was found in 60 cases. See Table 1 for a detailed summary of the correlation data.
Table 1
| Clinical characteristics | Total (n=379), n (%) |
|---|---|
| Age (years) | |
| <60 | 105 (27.7) |
| ≥60 | 274 (72.3) |
| Gender | |
| Female | 174 (45.9) |
| Male | 205 (54.1) |
| Stage | |
| I | 66 (17.4) |
| II | 152 (40.1) |
| III | 101 (26.6) |
| IV | 60 (15.8) |
| Stage-T | |
| T1 | 9 (2.4) |
| T2 | 66 (17.4) |
| T3 | 262 (69.1) |
| T4 | 42 (11.1) |
| Stage-N | |
| N0 | 227 (59.9) |
| N1 | 88 (23.2) |
| N2 | 64 (16.9) |
| Stage-M | |
| M0 | 319 (84.2) |
| M1 | 60 (15.8) |
TCGA, the Cancer Genome Atlas; Stage-T, the depth of tumor invasion; Stage-N, lymph node metastasis; Stage-M, distant metastasis.
Differential MFN2 expression
We downloaded information from 470 tumor samples and 41 normal (paracancerous) samples from TCGA. The Wilcoxon signed-rank test was performed to identify significant changes in MFN2 expression. MFN2 expression was considerably lower in colon cancer tissues than in paracancerous colon tissues in the original RNA expression study (P<0.001, Figure 1A). Further analysis revealed that the expression levels of MFN2 in tumor tissues were lower compared with matched paracancerous tissues (P<0.001, Figure 1B) in 41 paired samples. MFN2 expression was substantially linked with the clinical stage (stage IV vs. stage I, P=0.03, Figure 1C; stages I–III vs. stage IV, P=0.003, Figure 1D). MFN2 expression, as a categorical dependent variable, was predictive of clinicopathologic outcomes based on univariate analysis (see Table 2). In colon cancer patients, MFN2 expression was significantly associated with the AJCC stage [odds ratio (OR) =0.29 for stage IV vs. stage I, P=0.001], T-stage (OR =0.20 for T4vs. T1, P=0.033), and distant metastasis (OR =0.31 for M1vs. M0, P<0.001).
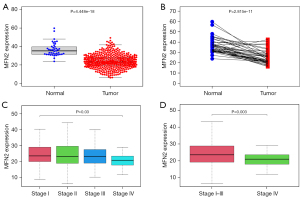
Table 2
| Clinical characteristics | Total | Odds ratio in MFN2 expression (95% CI) | P value |
|---|---|---|---|
| Stage (IV vs. I) | 126 | 0.29 (0.13–0.59) | 0.001 |
| Distant metastasis (M1 vs. M0) | 379 | 0.31 (0.16–0.55) | <0.001 |
| Age (≥60 vs.<60 years) | 379 | 1.07 (0.70–1.63) | 0.749 |
| Gender (male vs. female) | 379 | 0.78 (0.53–1.13) | 0.184 |
| T stage (T4 vs. T1) | 51 | 0.20 (0.04–0.81) | 0.033 |
| Lymph nodes (N2vs. N0) | 291 | 0.62 (0.37–1.02) | 0.063 |
MFN2, mitofusin 2; CI, confidence interval.
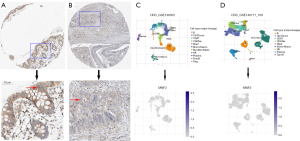
In addition, IHC findings were acquired and processed from the HPA to measure MFN2 expression at the protein level. MFN2 IHC staining was medium to high in colon normal tissues (Figure 2A), but MFN2 IHC staining was faint or not identified in CRC tumor tissues (Figure 2B). We employed single cell RNA seq data from the GSE139555 (14) and GSE146771 (15) datasets to explore MFN2 expression in a variety of cells, including colon cancer cells, immunological cells, stromal cells, and vascular endothelial cells. MFN2 expression was either low or non-existent in distinct clusters, according to differential gene analysis (Figure 2C,2D). These findings also revealed that MFN2 expression was low in colon cancer.
Kaplan-Meier survival analysis
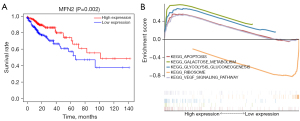
The colon cancer samples were divided into two groups based on the median MFN2 expression level: high- and low-MFN2 expression groups. We performed survival analysis to define the role of MFN2 in the prognosis of colon cancer. Based on Kaplan-Meier survival analysis, colon cancer patients with high MFN2 expression have a better prognosis compared to those with low MFN2 expression (P=0.002, Figure 3A).
Univariate and multivariate Cox analyses
We used univariate and multivariate Cox regression analyses to ascertain which characteristics, including age, sex, AJCC stage, TNM stage, and MFN2 expression level, were independent prognostic predictors for patients with colon cancer. Univariate Cox regression analysis showed that age [hazard ratio (HR) =1.03, 95% confidence interval (CI): 1.01–1.05, P=0.013], the AJCC stage (HR =2.34, 95% CI: 1.78–3.06, P<0.001), the depth of tumor invasion (HR =2.89, 95% CI: 1.81–4.59, P<0.001), distant metastasis (HR =4.83, 95% CI: 2.96–7.87, P<0.001), lymph node metastasis (HR =2.02, 95% CI: 1.54–2.66, P<0.001), and MFN2 expression (HR =0.94, 95% CI: 0.91–0.98, P=0.001) were associated with OS (Table 3). The multivariate Cox model indicated that age (HR =1.04, 95% CI: 1.01–1.06, P=0.001) and MFN2 expression (HR =0.95, 95% CI: 0.92–0.99, P=0.007) can be regarded as independent prognostic variables for colon cancer (Table 3).
Table 3
| Clinicopathologic variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age | 1.03 | 1.01–1.05 | 0.013 | 1.04 | 1.01–1.06 | 0.001 | |
| Gender | 1.11 | 0.70–1.78 | 0.653 | – | – | – | |
| Stage (AJCC) | 2.34 | 1.78–3.06 | 7.72E−10 | 1.68 | 0.76–3.71 | 0.20 | |
| T | 2.89 | 1.81–4.59 | 7.92E−06 | 1.53 | 0.88–2.67 | 0.13 | |
| M | 4.83 | 2.96–7.87 | 2.65E−10 | 1.36 | 0.47–3.97 | 0.58 | |
| N | 2.02 | 1.54–2.66 | 4.40E−07 | 1.23 | 0.75–2.01 | 0.41 | |
| MFN2 | 0.94 | 0.91–0.98 | 0.001 | 0.95 | 0.92–0.99 | 0.007 | |
AJCC, American Joint Committee on Cancer; Stage-T, the depth of tumor invasion; Stage-N, lymph node metastasis; Stage-M, distant metastasis; MFN2, mitofusin 2; HR, hazard ratio; OS, overall survival; CI, confidence interval.
GSEA
GSEA was performed using the high- and low-MFN2 expression datasets (false discovery rate P<0.05, nominal P<0.05) to identify putative signaling pathways involved in colon cancer, and we selected the most significantly enriched signaling pathways based on the NES. Using GSEA, we found that the high expression of MFN2 in colon cancer may be related to three metabolic signaling pathways, respectively KEGG GALACTOSE METABOLISM and KEGG GLYCOLYSIS GLUCONEOGENESIS. In addition, the high expression of MFN2 in colon cancer may be related to apoptosis and VEGF signaling pathways. However, GESA revealed only one signaling pathway KEGG RIBOSOME, which may be related to the low expression of MFN2 in colon cancer (Figure 3B, Table 4). These results indicate that changes in MFN2 expression may affect signaling pathways involved in the development and progression of colon carcinogenesis.
Table 4
| Name | ES | NES | NOM P value | FDR q-value |
|---|---|---|---|---|
| MFN2 high-expression | ||||
| KEGG_GALACTOSE_METABOLISM | 0.76 | 2.32 | <0.001 | <0.001 |
| KEGG_GLYCOLYSIS_GLUCONEOGENESIS | 0.68 | 2.21 | <0.001 | 0.002 |
| KEGG_VEGF_SIGNALING_PATHWAY | 0.54 | 2.11 | <0.001 | 0.006 |
| KEGG_APOPTOSIS | 0.53 | 1.98 | 0.002 | 0.010 |
| MFN2 low-expression | ||||
| KEGG_RIBOSOME | −0.83 | −1.81 | 0.016 | 0.137 |
MFN2, mitofusin 2; KEGG, Kyoto Encyclopedia of Genes and Genomes; ES, enrichment score; NES, normalized enrichment score; NOM, normal; FDR, false discovery rate.
Discussion
As early clinical symptoms are not obvious in CRC patients, approximately 35% of these patients present with metastatic disease at diagnosis (16). Moreover, over 50% of non-metastatic CRC patients ultimately develop metastatic disease. Aggressive treatment strategies, such as combining systemic chemotherapy with surgery, can sometimes cure patients with metastatic CRC. Molecular targeting with therapeutic drugs has been a landmark development in the treatment of metastatic CRC (17). Thousands of patients with metastatic CRC have received targeted therapies in clinical trials, and many patients experienced prolonged survival times or even a complete cure. However, the relative lack of biomarkers for metastatic CRC has slowed our progress in this area. As CRC-targeting therapies are currently limited, a better understanding of the pathogenesis and mechanisms of CRC is essential to develop targeted therapies.
Based on TCGA, we showed that tumor tissues had lower MFN2 expression levels than non-cancerous tissues (P<0.001), and there was lower expression in tumors at later tumor stages (stages I–III vs. stage IV, P=0.003). Besides, patients with low MFN2 expression had a worse prognosis than those with high MFN2 expression. Univariate and multivariate Cox analyses revealed that MFN2 expression may be an independent prognostic factor in colon cancer. A recent study reported low levels of MFN2 immunostaining in hepatocellular carcinoma (HCC) tissues compared to adjacent liver tissues (18). The lower expression of MFN2 was significantly connected to a poorer prognosis for patients with HCC. Overexpression of MFN2 induced apoptosis in HepG2 cells, stimulated Ca2+ release from the endoplasmic reticulum into the mitochondria, and increased intracellular reactive oxygen species (18,19). Furthermore, in HepG2 cells, the p53 protein is bound directly to the MFN2 promoter, indicating that MFN2 is a novel p53-inducible target gene (20). More recently, it was hypothesized that MFN2 may play a role in the onset and progression of HCC via a variety of mechanisms, including miRNAs; MFN2 is a direct target of miR-76 in HCC cells, and a miR-761 inhibitor upregulated MFN2, altering mitochondrial function and effectively inhibiting HCC cell growth and metastasis (21). These results suggest that MFN2 may be a potential therapeutic target for treating HCC. Downregulation of MFN2 is also linked to poor prognosis in breast cancer patients. Xu et al. used the CRISPR/Cas9 system to knock out MFN2 from MCF7 and A549 cells, stimulating tumor cell invasion in vitro and in vivo. Enhancing the mammalian target of rapamycin complex 2 (mTORC2)/Akt signaling pathway increases tumor cell proliferation and metastasis in MFN2-mutant cancer cells via an AktS437 phosphorylation-induced signal transduction pathway (22). In a separate investigation, MFN2 expression was considerably higher in 30 lung cancer samples compared to paired adjacent normal tissues (23). These contradictory findings suggest that the role of MFN2 in tumor growth is more complex than previously thought.
There are also contradictory reports about the involvement of MFN2 in the development of stomach or gastric cancer (GC). A recent study reported that patients with high MFN2 expression had significantly poorer OS and disease-free survival (DFS) than those with low or negative MFN2 expression. MFN2 overexpression was linked to the depth of invasion and TNM stage of GC, and MFN2 knockdown prevented the migration and invasion of GC cells (9). Zhang et al. found lower MFN2 expression in tumor tissue than in normal tissue and that the expression level was inversely connected with tumor size, the clinical stage, lymph node metastasis, and distant metastasis. MFN2 expression had no significant impact on OS or DFS, but MFN2 overexpression reduced GC cell proliferation, clonogenicity in vitro, and tumor growth in vivo, which were associated with the downregulation of MMP-2 and -9 (24). These inconsistent and conflicting results suggest that the role of MFN2 in GC is also more complicated than initially thought.
As a fusion protein of mitochondria, the specific mechanism of MFN2 involved in the occurrence of CRC is still unclear. Mitochondria have long been regarded as the cell’s “powerhouse” as they produce more than 90% of ATP in aerobic settings via oxidative phosphorylation. To maintain optimal mitochondrial activities, each cell maintains a precise balance between fusion and fission. Mitofusin-1 (MFN1), mitofusin-2 (MFN2), and optic atrophy 1 (OPA1) are the three primary dynamin-related proteins that mediate mitochondrial fusion in mammals (25). Abnormal MFN2 function can cause mitochondrial fusion dysfunction, leading to a variety of diseases, such as diabetes, obesity, cardiovascular and cerebrovascular diseases, neurodegenerative diseases, malignant tumors and so on (8,26,27). The increased glucose intake and increased glycolytic rates suggest that metabolic changes help tumor cells grow faster (28). MFN2 reduces cancer cell growth by interacting with pyruvate kinase 2 (PKM2) via the N-terminus and restricting metabolic flow to aerobic glycolysis; phosphorylation of Mfn-2 strengthens this connection (29). The loss of MFN2 activity induces metabolic changes in mitochondria, including decreased mitochondrial membrane potential, cellular oxygen consumption, and substrate oxidation. Our study also found that high expression of MFN2 in colon cancer may be involved in KEGG GALACTOSE METABOLISM and KEGG GLYCOLYSIS GLUCONEOGENESIS pathway, suggesting that the development of colon cancer may be related to the abnormal mitochondrial energy metabolism caused by MFN2.
In the early stages of intrinsic apoptosis, mitochondria are the principal target (30). Dynamic changes in the mitochondrial location of numerous proteins, including MFN2 and Drp1, as well as the apoptosis regulators Bcl-2 associated X (BAX) and Bcl-2 antagonist/killer (BAK), cause mitochondrial fragmentation during apoptosis (31). Overexpression of either MFN1 or MFN2 can also delay apoptosis by delaying the activation of downstream caspases and apoptosis (32). MFN2 location on the outer mitochondrial membrane is known to be mediated by BAX, while BAK binds to both MFN1 and MFN2 (33). Several investigations have shown that MFN2-induced apoptosis is most likely mediated through the PI3K-Akt pathway (34,35). Our GSEA showed that high expression of MFN2 may promote apoptosis in colon cancer cells, but the specific inhibition needs to be further elucidated. Vascular endothelial growth factor (VEGF) upregulates MFN2 expression in human umbilical vein endothelial cells, and knocking down MFN2 reduces VEGF-induced human umbilical vein endothelial cell migration and differentiation, indicating that MFN2/VEGF connection in endothelial cells plays a role in angiogenesis (36). Another study showed that overexpressed MFN2 has an inhibitory effect on pancreatic cancer, which may be related to the VEGF signaling pathway (37). Our analysis also found that high expression of MFN2 may be involved in the VEGF signaling pathway in colon cancer. VEGF plays a key regulatory role in the growth and invasion of CRC. The VEGF inhibitor bevacizumab provides a significant survival benefit for advanced CRC treatment. Clarification of the relationship between MFN2 and VEGF in colon cancer cells is likely to bring new ideas for targeted therapy of CRC.
Analysis of data from TCGA for rectal cancer (data not shown) did not produce similar results, suggesting that MFN2 may have different functions depending on the site. Due to data heterogeneity or software issues, our results may be biased or erroneous. More data are needed, and further experiments and clinical validation should be conducted to confirm the link between MFN2 and colon cancer.
Conclusions
To the best of our knowledge, this is the first study of the role of MFN2 in colon cancer. Low MFN2 expression implies a poor prognosis and is an independent risk factor for colon cancer prognosis. We also showed that MFN2 may be a potential prognostic biomarker and therapeutic target for colon cancer. A comprehensive evaluation of the mode of action of MFN2 may provide insight into the role of MFN2 in the genesis of colon cancer and lead to targeted molecular therapies.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-589/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-589/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-589/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Jung G, Hernández-Illán E, Moreira L, et al. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol 2020;17:111-30. [Crossref] [PubMed]
- Cheng X, Xu X, Chen D, et al. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother 2019;110:473-81. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Chen KH, Guo X, Ma D, et al. Dysregulation of HSG triggers vascular proliferative disorders. Nat Cell Biol 2004;6:872-83. [Crossref] [PubMed]
- Rocha AG, Franco A, Krezel AM, et al. MFN2 agonists reverse mitochondrial defects in preclinical models of Charcot-Marie-Tooth disease type 2A. Science 2018;360:336-41. [Crossref] [PubMed]
- Chandhok G, Lazarou M, Neumann B. Structure, function, and regulation of mitofusin-2 in health and disease. Biol Rev Camb Philos Soc 2018;93:933-49. [Crossref] [PubMed]
- Fang CL, Sun DP, Chen HK, et al. Overexpression of Mitochondrial GTPase MFN2 Represents a Negative Prognostic Marker in Human Gastric Cancer and Its Inhibition Exerts Anti-Cancer Effects. J Cancer 2017;8:1153-61. [Crossref] [PubMed]
- Xue R, Meng Q, Lu D, et al. Mitofusin2 Induces Cell Autophagy of Pancreatic Cancer through Inhibiting the PI3K/Akt/mTOR Signaling Pathway. Oxid Med Cell Longev 2018;2018:2798070. [Crossref] [PubMed]
- Allegra A, Innao V, Allegra AG, et al. Relationship between mitofusin 2 and cancer. Adv Protein Chem Struct Biol 2019;116:209-36. [Crossref] [PubMed]
- Cheng X, Zhou D, Wei J, et al. Cell-cycle arrest at G2/M and proliferation inhibition by adenovirus-expressed mitofusin-2 gene in human colorectal cancer cell lines. Neoplasma 2013;60:620-6. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Wu TD, Madireddi S, de Almeida PE, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 2020;579:274-8. [Crossref] [PubMed]
- Zhang L, Li Z, Skrzypczynska KM, et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 2020;181:442-459.e29. [Crossref] [PubMed]
- Zacharakis M, Xynos ID, Lazaris A, et al. Predictors of survival in stage IV metastatic colorectal cancer. Anticancer Res 2010;30:653-60. [PubMed]
- Fakih MG. Metastatic colorectal cancer: current state and future directions. J Clin Oncol 2015;33:1809-24. [Crossref] [PubMed]
- Wang W, Xie Q, Zhou X, et al. Mitofusin-2 triggers mitochondria Ca2+ influx from the endoplasmic reticulum to induce apoptosis in hepatocellular carcinoma cells. Cancer Lett 2015;358:47-58. [Crossref] [PubMed]
- Wang L, Yang X, Shen Y. Molecular mechanism of mitochondrial calcium uptake. Cell Mol Life Sci 2015;72:1489-98. [Crossref] [PubMed]
- Wang W, Cheng X, Lu J, et al. Mitofusin-2 is a novel direct target of p53. Biochem Biophys Res Commun 2010;400:587-92. [Crossref] [PubMed]
- Zhou X, Zhang L, Zheng B, et al. MicroRNA-761 is upregulated in hepatocellular carcinoma and regulates tumorigenesis by targeting Mitofusin-2. Cancer Sci 2016;107:424-32. [Crossref] [PubMed]
- Xu K, Chen G, Li X, et al. MFN2 suppresses cancer progression through inhibition of mTORC2/Akt signaling. Sci Rep 2017;7:41718. [Crossref] [PubMed]
- Lou Y, Li R, Liu J, et al. Mitofusin-2 over-expresses and leads to dysregulation of cell cycle and cell invasion in lung adenocarcinoma. Med Oncol 2015;32:132. [Crossref] [PubMed]
- Zhang GE, Jin HL, Lin XK, et al. Anti-tumor effects of Mfn2 in gastric cancer. Int J Mol Sci 2013;14:13005-21. [Crossref] [PubMed]
- Rodrigues T, Ferraz LS. Therapeutic potential of targeting mitochondrial dynamics in cancer. Biochem Pharmacol 2020;182:114282. [Crossref] [PubMed]
- Kim IS, Silwal P, Jo EK. Mitofusin 2, a key coordinator between mitochondrial dynamics and innate immunity. Virulence 2021;12:2273-84. [Crossref] [PubMed]
- Cormio A, Busetto GM, Musicco C, et al. Mitofusin-2 Down-Regulation Predicts Progression of Non-Muscle Invasive Bladder Cancer. Diagnostics (Basel) 2021;11:1500. [Crossref] [PubMed]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011;27:441-64. [Crossref] [PubMed]
- Li T, Han J, Jia L, et al. PKM2 coordinates glycolysis with mitochondrial fusion and oxidative phosphorylation. Protein Cell 2019;10:583-94. [Crossref] [PubMed]
- Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell 2011;21:92-101. [Crossref] [PubMed]
- van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol 2013;5:a011072. [Crossref] [PubMed]
- Srinivasan S, Guha M, Kashina A, et al. Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim Biophys Acta Bioenerg 2017;1858:602-14. [Crossref] [PubMed]
- Brooks C, Wei Q, Feng L, et al. Bak regulates mitochondrial morphology and pathology during apoptosis by interacting with mitofusins. Proc Natl Acad Sci U S A 2007;104:11649-54. [Crossref] [PubMed]
- Datta SR, Dudek H, Tao X, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997;91:231-41. [Crossref] [PubMed]
- Kim NH, Kim K, Park WS, et al. PKB/Akt inhibits ceramide-induced apoptosis in neuroblastoma cells by blocking apoptosis-inducing factor (AIF) translocation. J Cell Biochem 2007;102:1160-70. [Crossref] [PubMed]
- Lugus JJ, Ngoh GA, Bachschmid MM, et al. Mitofusins are required for angiogenic function and modulate different signaling pathways in cultured endothelial cells. J Mol Cell Cardiol 2011;51:885-93. [Crossref] [PubMed]
- Lin Z, Lin X, Chen J, et al. Mitofusin-2 is a novel anti-angiogenic factor in pancreatic cancer. J Gastrointest Oncol 2021;12:484-95. [Crossref] [PubMed]


