
Development and validation of a novel ubiquitination-related gene prognostic signature based on tumor microenvironment for colon cancer
Introduction
Colon cancer (CC) is one of the most common cancers with high morbidity in the world (1), posing a severe threat to human health. According to GLOBOCAN 2021 estimates of cancer incidence and mortality, CC has the highest incidence in Western and Northern Europe, with a slightly higher incidence in men than in women (2). Most cases are diagnosed at a late stage, leading to a poor prognosis for this cancer (3), and the 5-year survival rate for metastatic CC patients is below 15% (4). Chemotherapy is one of the most frequently used therapies for CC (5), but its drug resistance and severe side effects limit its further clinical application (6). Therefore, more CC biomarkers with high efficiency and sensitivity need to be explored.
Ubiquitination, the post-translational addition of ubiquitin proteins to target proteins, controls various cellular processes by altering protein stability, localization, activity, or interactions (7). Since ubiquitination regulates many signaling pathways and is involved in many biological processes (8), including substantial modifications of proteins that regulate cellular functions, aberrant ubiquitination may lead to cancer (9). In breast and ovarian cancer, CUL3-mediated ubiquitination and degradation of BECN1 could promote the proliferation of tumor cells, and the expression of CUL3 was related to poor prognosis in patients (10). Huang et al. suggested that UBE2O is an attractive radiosensitization target for lung cancer treatment. In addition, several ubiquitinating enzymes are associated with the prognosis of CC (11). NEDD4, a core member of the NEDD4 E3 ubiquitin ligase family, is abundantly expressed in CC tissues and cells. Its overexpression promotes xenograft tumor growth, metastasis, and tumorigenicity of primary CC in mouse models (12). Cul4A, an essential component of the E3 ubiquitin ligase family, of which expression is positively correlated with the prognosis of colorectal cancer (13). Evaluation of CUL4A and mutant TP53 expression will help predict the prognosis of CC (14). In short, tumor ubiquitination may be critical in determining the prognosis and oncologic treatment of CC patients.
The role of the ubiquitin-proteasome system in tumorigenesis is not only related to tumor metabolic regulation (mTORC1/AMPK/AKT) but also to the tumor microenvironment (TME) regulation (TLR/RLR/STING) (15). TME is recognized as a hotbed of tumor cells and consists of cell types such as tumor cells, endothelial cells, stromal cells, and immune cells and is a critical factor in multiple stages of tumor progression (16). The role of ubiquitination playing in TME has received widespread attention. Guo et al. demonstrated that E3 ligase NEDD4 mediates ubiquitination and degradation of GITR and suppresses T-cell-mediated-killings on melanoma cells (17). Zhang et al. suggested activating the USP7/hnRNPA1 axis promotes miR-522 secretion by cancer-associated fibroblasts (CAFs) and ultimately reduces chemosensitivity in gastric cancer (18). Besides, there is growing evidence that TME can determine abnormalities in tissue function and play an essential role in developing more advanced and refractory malignancies (19). Considerable studies have shown that tumor-associated stromal cells are closely associated with T-cell suppression (20), CC cell metastasis (21-24), and drug resistance (25) in CC. Considering the importance of ubiquitination and TME in tumor progression and prognosis and the tight relationship between them, we believe it is crucial to develop and validate a CC prognostic model related to ubiquitination and assess its correlation with TME. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-607/rc).
Methods
Data sources and preprocessing
A total of 2 CC cohorts, TCGA-COAD and GSE17538, were recruited from The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO). We downloaded all available data on somatic mutations, transcriptome profiles, microsatellite instability status (MSI), and clinical information of CC from the TCGA database. A total of 399 samples with somatic mutation data were analyzed to show the mutation profile of CC. Then, survival analysis was performed after obtaining clinical data of CC patients from the TCGA database (n=452) and the GSE17538 dataset (n=238). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of URGs
A total of 247 URGs with relevance score >10 were obtained from the GeneCards database. Then, ten URGs sets with M12685, M15079, M15645, M23353, M23636, M24394, M24634, M27401, M27742, and M42893 were downloaded from GSEA. The 247 genes from GeneCards were crossed with each of the ten sets from the GSEA database. Finally, 56 genes related to ubiquitination were selected for further analysis.
Unsupervised Clustering of 56 URGs
Consensus clustering was performed by the R package “ConsensusClusterPlus” to classify CC patients into two subgroups based on the expression differences of 56 URGs (26). The chosen clustering algorithm was K-means and the distance metric was Euclidean. In addition, we performed the above steps 50 times to ensure the stability of the classification.
Construction and validation of the prediction model
To further screen out URGs with high predictive value, we performed Cox regression analysis in the combined of TCGA and GSE17538, which assess the relationship between URGs and survival status. Nine URGs were firstly study identified for further analysis with adjusted P<0.005. Least absolute shrinkage and selection operator (LASSO) regression analysis was performed by the R package “glmnet” to identify prognostically relevant URGs and to model the risk. Finally, seven genes were selected as the best to construct the risk score. The risk scores were calculated as following: (X: coefficients, Y: gene expression level). Next, data from the TCGA database was used as the training group. It was divided into high-risk and low-risk groups based on the median risk score, and Kaplan-Meier survival analysis was plotted. In addition, receiver operating characteristic (ROC) curves for 1, 3, and 6 years were generated in R software to calculate the area under the ROC curve (AUC) values for each predictor model, further to assess the efficiency and accuracy of the model. Data from GSE17538 was treated as the test group, and the same procedure was performed to validate the prognostic models in this group. P<0.05 was considered to be statistically significant.
Construction protein-protein interaction (PPI) network
PPI information for 7 URGs was obtained from the STRING database; the highest confidence level of 0.400 was selected (version 11.5, https://string-db.org/, accessed data: 2022/5/24).
Expression of 7 URGs in the single cell-type clusters identified
The expression of 7URGs in the single cell-type clusters identified were obtained from the Human Protein Atlas (version 21.0, https://www.proteinatlas.org/, accessed data: 2022/5/24).
GSVA between high-risk and low-risk groups in the training group
To further explore the differences in biological pathway activation status between the high-risk and low-risk groups in the training group, GSVA enrichment analysis was performed using the R package “GSVA” (27). The gene set “c2.cp.kegg.v7.2.symbols” was downloaded from the MSigDB database (27). Adjusted P value <0.05 was considered statistically significant.
Estimation of immune cell infiltration
The single-sample gene set enrichment analysis (ssGSEA) algorithm was used to quantify the relative infiltration of 28 immune cell types within the TME. The enrichment fraction was calculated using ssGSEA represents the abundance of immune cells within the TME in each sample. The featured gene panels for each distinct immune cell were obtained from the study of Charoentong et al. (28).
Calculation of TME scores
The ratio of immune/matrix components in the TME of each tumor sample was analyzed by R package “estimate”. The immune score, matrix score, and estimated score reflect the corresponding ratios of the immune component, matrix component, and the sum of the two in the TME. Higher scores represent a more excellent ratio of immune/stromal components in TME.
Tumor mutation burden (TMB) analysis
Somatic mutation data were processed and analyzed using R package “maftools” to explore the mutational landscape of CC. TMB was defined as the sum of somatic mutations, insertional deletion mutations, coding, and base substitutions per million bases.
Statistical analysis
R software (version 4.1.2), SPSS (version 23.0), and R studio (version 1.1.463) were used to perform statistical analyses. Single-factor analysis was utilized to compare the differential expression of genes. Person chi-square test was used to assess categorical variables. Kaplan-Meier method combined with logit test was used to analyze the overall survival (OS) of CC patients. Univariate and multivariate Cox regression was used to assess the independent prognostic value of the risk model. Wilcoxon test was used to analyze immune cell infiltration and immune pathways.
Results
Identifying prognostic-related URGs that were associated with CC
A total of 56 URGs were screened as described in the Materials and Methods section. Among the 56 URGs, five genes, NEDD4L, PRKN, UBB, UBE2E2, and WWP2, were significantly downregulated in tumor samples, while ARIH2, BTRC, CDC34, CUL1, NEDD4, OTUB1, RBX1, RNF168, RNF8, RPS27A, SKP2, STUB1, TRAF6, UBA1, UBA52, UBA6, UBE2B, UBE2C, UBE2D1, UBE2D2, UBE2D3, UBE2D4, UBE2E3, UBE2G1, UBE2G2, UBE2H, UBE2J2, UBE2N, UBE2R2, UBE2S, UBE2T, UBE2V1, UBE2W, UBE3C, UBR5, UCHL3, USP5, USP7, USP8, USP9X, VCP, were remarkably upregulated in tumor samples (Figure 1A), indicating the potential role of these cryptic URGs in CC development.
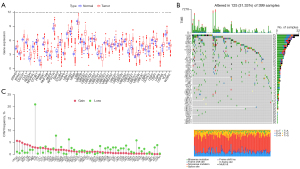
Genetic alterations of URGs in CC
We examined the incidence of somatic mutations and the frequency of somatic copy number variations (CNVs) for these sample genes. The mutation details for each CC sample are shown in the waterfall plot. Among the 399 samples, USP9X had the highest mutation frequency, but only 8%, followed by UBR5, UBA6, USP7, UBE3A, NEDD4, CUL1, UBE3C, CUL3, NEDD4L, USP8, USP5, VCP, ITCH, BTRC, RNF168, RNF8, STUB1, UBA1, WWP2, TRAF6, UBC, UBE2A, UBE2E2, UBE2K, UBE2L3, UBE2R2, ARIH2, CDC34, OTUB1, UBB, UBE2J2, AMFR, SKP2, UBE2B, UBE2E1, UBE2G1, UBE2T, SKP1, UBA52, UBE2E3, UBE2G2, and UBE2W, while the rest did not show mutations (Figure 1B). In addition, we found that PRKN gene had the highest CNV, followed by UBE2J2, BTRC, UBE3A, and other genes. The CNVs of all genes, except PRKN, were below 10%, implying that they may not play an important role in the biology of CC (Figure 1C).
Biological characteristics and prognosis of each ubiquitination-associated subtype
The R package “ConsensusClusterPlus” was used to divide patients in the combined TCGA and GSE17538 cohorts into two subgroups based on the expression levels of the 56 URGs. Finally, two different clusters were identified using the unsupervised clustering method: 180 cases in cluster A and 507 cases in cluster B (Figure 2A). Principal component analysis revealed the existence of distinct transcriptome profiles between the two clusters (Figure 2B). Prognostic analysis of the two clusters showed no significant difference in survival between the two clusters (Figure 2C).
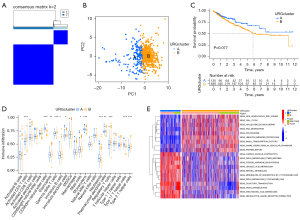
GSVA was performed to explore the biological behavior of the two clusters. As shown in Figure 2D, cardiac muscle contraction, drug metabolism, steroid hormone biosynthesis, linoleic acid metabolism, and retinol metabolism were significantly enriched in cluster A. Cluster B displayed enriched pathways such as non-homologous end-joining, cell cycle, Nucleotide excision repair, RNA degradation, and spliceosome. Infiltrating lymphocytes within TME were subsequently analyzed using the ssGSEA algorithm (Figure 2E). Although cluster B showed worse survival results, immune cell infiltrates such as activated CD4+ T cells, eosinophilna, γδ T cells, immature B cells and dendritic cells, mast cells, plasmacytoid dendritic cells, regulatory T cells, and type 2 T helper cells were significantly enriched. In contrast, only CD56bright natural killer cells, monocytes, neutrophils and type 17 T helper cells were significantly enriched in cluster A.
Identification of survival-related URGs in CC
To explore the possible relationship between URGs and OS in CC patients, we used univariate Cox analysis to select survival-related URGs in the combined TCGA and GSE17538 cohorts as an initial screen. A total of 9 OS-associated URGs (adjusted P value <0.005) were identified. Information on Gene Symbol, HR, 95% CI, and P value are shown in Table 1. Next, LASSO Cox regression analysis was used to identified 7 survival-related genes (UBE2B, UBE2E2, UBE2G2, UBE2K, USP7, USP8, USP9X) to construct risk model for prognosis according to the λ=0.001 (Figure 3A,3B), and the corresponding coefficients were obtained (as shown in Table 2). The risk score formula was as fellow: risk score = (0.003643 * UBE2B exp.) + (0.200524 * UBE2E2 exp.) + (0.801445 * UBE2G2 exp.) + (−0.54965 * UBE2K exp.) + (−0.45943 * USP7 exp.) + (0.241099 * USP8 exp.) + (−0.0726 * USP9X exp.). These selected genes were classified as risky (UBE2B, UBE2E2, USP8, UBE2G2), where HR >1 was associated with a poorer prognosis, and protective (USP7, UBE2K, USP9X), where HR <1 was associated with a better prognosis (Table 2). The analysis of PPI network construction can be used to examine the interaction between the seven genes involved in the risk modeling (Figure 3C). And the correlations between the 7-URG were displayed in the network diagram (Figure 3D). The 7-URG expression in the single cell type clusters identified was shown in Figure S1. USP8, USP9X, and UBE2K have low cell-type specificity. At the same time, USP7 is enhanced in early spermatids, UBE2B is enhanced in syncytiotrophoblasts, UBE2G2 is enhanced in late spermatids, and UBE2E2 is enhanced in oligodendrocytes, excitatory neurons. Besides, the expression of 7-URG in the single cell-type clusters identified in the colon can be seen in Figure S2.
Table 1
| Gene symbol | Overall survival | |||
|---|---|---|---|---|
| HR | 95% CI | P value | Adjusted P value | |
| BTRC | 1.82359 | 1.148597–2.895256 | 0.010854 | 0.004171 |
| UBE2B | 1.255957 | 0.904488–1.744001 | 0.173633 | 6.32E-05 |
| UBE2E2 | 1.275731 | 1.078707–1.508741 | 0.004439 | 0.000363 |
| UBE2G2 | 1.599679 | 1.092623–2.342045 | 0.015719 | 0.004014 |
| UBE2K | 0.712834 | 0.488272–1.040676 | 0.079527 | 0.001678 |
| UBR5 | 1.216318 | 0.933420–1.584954 | 0.1471 | 0.002319 |
| USP7 | 0.649282 | 0.442717–0.952227 | 0.02707 | 0.001934 |
| USP8 | 1.419101 | 1.026131–1.962563 | 0.034353 | 0.00092 |
| USP9X | 0.896795 | 0.690060–1.165466 | 0.415236 | 0.002969 |
URGs, ubiquitination-related genes; HR, hazard ratio; CI, confidence interval.
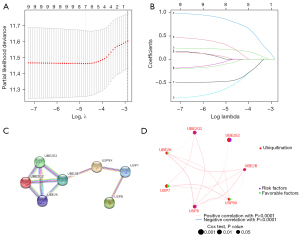
Table 2
| Gene symbol | Overall survival | ||||
|---|---|---|---|---|---|
| Coef | HR | 95% CI | P value | Adjusted P value | |
| USP7 | −0.45943 | 0.649282 | 0.442717–0.952227 | 0.02707 | 0.001934 |
| UBE2K | −0.54965 | 0.712834 | 0.488272–1.040676 | 0.079527 | 0.001678 |
| USP9X | −0.0726 | 0.896795 | 0.69006–1.165466 | 0.415236 | 0.002969 |
| UBE2B | 0.003643 | 1.255957 | 0.904488–1.744001 | 0.173633 | 6.32E-05 |
| UBE2E2 | 0.200524 | 1.275731 | 1.078707–1.508741 | 0.004439 | 0.000363 |
| USP8 | 0.241099 | 1.419101 | 1.026131–1.962563 | 0.034353 | 0.00092 |
| UBE2G2 | 0.801445 | 1.599679 | 1.092623–2.342045 | 0.015719 | 0.004014 |
URGs, ubiquitination-related genes; Coef, coefficient; HR, hazard ratio; CI, confidence interval.
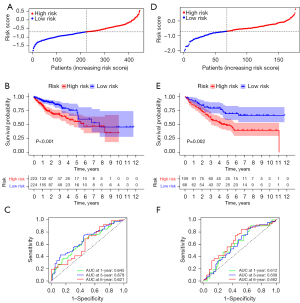
Subsequently, we used 452 CC patients from the TCGA database as the training group and 238 patients from GSE17538 as the test group. Patients in the training group were divided into high-risk and low-risk groups based on their risk scores, and the median risk score was set as the cut-off point (Figure 4A). As the risk score increased, the number of deaths increased, and survival time decreased. The Kaplan-Meier curve showed that CC patients with high-risk had a worse prognosis than those with low-risk (Figure 4B). In addition, the ROC curve demonstrated the predictive power of the risk score on prognosis; the AUCs were 0.645, 0.678, and 0.621 for 1, 3, and 6 years, respectively (Figure 4C). We used GSE17538 for validation and repeated the above analysis. The same results were obtained (Figure 4D,4E). The risk score had good prognostic predictive power with AUCs of 0.612, 0.609, and 0.662 at 1, 3, and 6 years, respectively (Figure 4F).
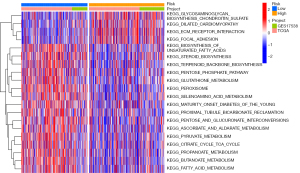
Differential biobehavior of high- and low-risk subgroups identified by GSVA analysis
Inspired by the ability of the seven-gene signature to predict prognosis in the training and test groups, we further performed GSVA analysis to explore differences in biological pathway activation status between high-risk and low-risk groups, aiming to find a more comprehensive explanation for prognostic differences between high-risk and low-risk groups (Figure 5). Heat maps were used to visualize the top 20 biological processes with significant differences, ranked according to P value from smallest to largest. We found a significant enrichment of activation pathways for carcinogenesis in the high-risk group, such as glycan metabolism, Dilated cardiomyopathy, ECM-receptor interaction, and cytoskeletal regulation, which may provide some perspective on the reasons for the poor prognosis in the high-risk group.
The relevance of TME to URGs expression
Different levels of immune infiltration may explain to some extent why patients with the same histological type of cancer may have different clinical outcomes. We explored the correlation between 7 URGs and the level of immune infiltration, aiming to reveal the potential mechanisms by which seven genetic features affect CC prognosis. Interestingly, violin plot analysis showed that the stromal cell and immune cell content and the combined content were higher in the high-risk group than in the low-risk group (Figure 6A). The level of immune cell infiltration in the high-risk group, including B cells, activated CD4+ and CD8+ T cells, dendritic cells, Eosinophilna were higher than those in the low-risk group (P<0.05), while the low-risk group was enriched with follicular helper T cells (Tfh cells) and resting NK cells (P<0.05) (Figure 6B). Recent studies have shown that although some tumor tissues possess a large number of immune cells, these immune cells, such as T-cells, are trapped in the surrounding stroma (29-32). Therefore, the activation of stromal cells in the TME is considered to be immunosuppressive (33,34). Apparently, the high-risk group reflects a TME filled with a large number of immunosuppressive cells and stromal cells, which is consistent with the poor prognosis. TIMER was then applied to assess the correlation between the expression levels of the seven URGs and the levels of tumor purity and immune cell infiltration (Figure 6C). Besides, we found that resting dendritic cells and CD8+ T cells were negatively correlated with risk scores, while neutrophils and gamma delta T cells were positively correlated with risk scores (Figure 6D-6G). The results showed that gene expression has an extremely complex and variable impact on the microscopic representation of immune infiltration, reflecting the heterogeneity and complexity of the immune microenvironment.

Interestingly, as protective types, UBE2K, USP7, and USP9X were significantly negatively correlated with T-cell infiltration and positively correlated with macrophage infiltration. Most of the risky genes showed a trend of positive correlation with macrophage infiltration and negative correlation with B cells. These findings suggest that the seven-gene profile may play an essential role in the formation and layout of CC immune infiltrates. Then, we analyzed in detail the correlation between risk scores and resting dendritic cells, neutrophils, CD8+ T cells, and gamma delta T cells. The risk score was negatively correlated with resting dendritic cells and CD8+ T cells and was positively correlated with neutrophils and gamma delta T cells.
Differences in tumor mutation profiles and MSI status between high- and low-risk groups
Mutation details for each CC sample in the high and low-risk groups are shown separately in the waterfall plot (Figure 7A,7B), in which we can analyze the different mutation types of each gene involved in CC progression. We next analyzed the differences in TMB between the high- and low-risk groups separately and found that the TMB in the low-risk group was smaller than that in the high-risk group (P<0.001) (Figure 7C). In addition, we obtained MSI data from the TCGA database. Based on the MSI classification method, a total of 78 samples were classified as MSI-High (MSI-H), and 344 samples were classified as MSI-low (MSI-L)/microsatellite stable (MSS). Samples in the low-risk group consisted of 70% by MSS, 17% by MSI-L, and 14% by MSI-H. The high-risk group samples consisted of 57% by MSS, 18% by MSI-L, and 24% by MSI-H. The MSS samples had a lower risk score than MSI-H (P=0.0014) (Figure 7D,7E).
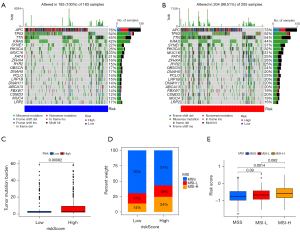
Current studies suggest that CRCs with MSI-H have a better prognosis than MSS, which seems contradictory to our results showing a higher risk score for MSI-H than the MSS group (35,36). However, some studies also have highlighted that MSI-H patients have a better sensitivity to 5-fluorouracil than MSS (37-39), making the prognosis of MSI-H patients under 5-fluorouracil therapy potentially favorable. In our study, the treatment regimen of TCGA-COAD patients was unknown. Furthermore, the data of GSE17538 contained a large number of patients without adjuvant chemotherapy (40), making the prognosis of MSI-H patients in our study not necessarily better than that of MSS patients. Therefore, the risk score of MSI may be higher than that of MSS. Popat et al. found that although some studies reported high survival rates for patients with MSI, estimates of the prognostic value of MSI vary widely between studies (41). The impact of MSI on the prognosis of patients with CC still needs to be further explored.
Discussion
Ubiquitination has recently received wide attention due to its significance in normal biological processes (42). For example, Huang et al. found that UBE2O promotes tumorigenesis and radiation tolerance via facilitating Mxi1 ubiquitination and degradation, suggesting that UBE2O is an attractive radiosensitization target for lung cancer treatment (11). Li et al. found that the degradation of PDCD4, an essential ubiquitinated substrate of SKP2, can promote tumorigenesis and radiation tolerance in breast cancer (43). In addition, NEDD4 was shown to trigger FOXA1 ubiquitination and promote CC progression (12).
However, a single gene signature is susceptible to multiple factors, making it difficult to be a reliable prognostic marker. Our study screened 56 URGs from the GeneCards and GSEA databases. Then we obtained 452 expression data and clinical profiles from the TCGA database. Univariate and multivariate Cox regression analyses were performed to determine the prognostic value of the 7 URGs (USP7, UBE2K, USP9X, UBE2B, UBE2E2, USP8, and UBE2G2) for CC patients. Among the seven genes, USP7, USP9X, and USP8 are all deubiquitinating enzymes (DUBs). Quite a few studies have shown that USP7 promotes tumor cell development, progression, and metastasis by deubiquitinating the p53 negative regulatory protein MDM2, which decreases intracellular levels of p53 (44-47). However, USP7 also prevents genetic alterations in various ways independent of p53. It promotes telomere maintenance, repairs broken double-stranded DNA, and regulates DNA damage checkpoint 1-mediated protein stability, which may reduce genomic instability and gene amplification, leading to tumorigenesis (48,49). These functions probably make it a favorable factor in our results (50). The crystal structure of USP9X was shown to be close to USP7, with a canonical USP-fold comprised of fingers, palm, and thumb subdomains, as well as an unusual β-hairpin insertion that may have significant effects on its function. Therefore, we speculate that the structural similarity between USP9X and USP7 may have led to their functional similarity, ultimately making them together as favorable factors. Although USP8 is a DUB, it was classified as a risky factor in our study, which may be related to its remodeling of TME. It has been suggested that inhibition of USP8 increases PD-L1 protein abundance and activates NF-κB signaling to trigger innate immune responses and MHC-I expression. Combining USP8 inhibitors with PD-1/PD-L1 blockers inhibited tumor growth and improved survival in CC mouse model (51). Conversely, high levels of USP8 may lead to T cell dysfunction, consistent with our results that USP8 is a risky factor and its expression is negatively correlated with CD8+ T cells (51,52). UBE2B, UBE2G2, UBE2E2 and UBE2K are all E2 ubiquitin-conjugating enzymes and E2s are classified into four different types (53). Studies have suggested that downregulation of UBE2B, also known as Rad6B, attenuates the expression of cancer stem cell markers (54-57). Rad6B protein promotes proliferation and invasion of normal human hepatocytes and has been associated with tumorigenesis and platinum resistance in breast cancer (58-60) and ovarian cancer (61). Meanwhile, Menezes et al. identified deleterious in Blastic Plasmacytoid Dendritic Cell Neoplasm UBE2G2 mutation (62). These findings are consistent with our results that UBE2B and UBE2G2 are risky factors. UBE2E2 is a class II E2 whose effects in tumors are currently unknown. UBE2K, a class III E2, is a favorable factor containing a unique C-terminal. It has been demonstrated that UBE2K is involved in mediating polyglutamine aggregate formation and cell death (63). This indicates the importance of ubiquitination for cellular clearance or storage of toxic proteins beyond the proteasome itself, possibly qualifying it as a favorable factor. Subsequent survival analysis showed that the high-risk group was associated with a poorer prognosis, which was validated by GSE17538.
In addition, through GSVA analysis, we found that the high-risk group was significantly enriched for oncogenic activation pathways such as Dilated cardiomyopathy, Glycan metabolism, ECM-receptor interaction and Cytoskeletal regulation, which provides a more comprehensive explanation for its poor prognosis. ARC is an anti-apoptotic protein abundant in cardiomyocytes and plays an important role in mediating apoptosis in dilated cardiomyopathy (64). Foo et al. found that ARC degradation is dependent on the p53-induced ubiquitin E3 ligase MDM2 (65), which USP7 can deubiquitinate (66). Therefore, we suggest that USP7 is involved in the development of dilated cardiomyopathy through the p53/MDM2 pathway. Although the relationship between Glycan metabolism (67), ECM-receptor interaction (68-71), Cytoskeletal regulation (72), and 7-URG has not been investigated, ubiquitination is involved in regulating cellular functions by these three pathways. Meanwhile, increasing studies have suggested that these three pathways are closely related to tumor growth and metastasis (73-75), so we speculate that 7-URG is related to the three pathways regulating tumor cell function. These findings suggest that the 7-URG risk model has a powerful ability to predict the prognosis of CC patients and may have implications for clinical treatment decisions.
Few existing studies have focused on the relationship between tumor ubiquitination and the TME (15,76). Our results showed that infiltration of both stromal cells and immune cells (especially immunosuppressive cells) was significantly higher in the high-risk group than in the low-risk group. Furthermore, we uncovered a correlation between each immune cell and 7-URG expression. Notably, we found CD8+ T cells correlated with both 7-URG expression, especially with three risky genes, UBE2B, UBE2E2, and USP8, in a significant negative correlation (P<0.001). Then, we found that CD8+ T cells were negatively correlated with risk score (P=0.014). All these results are consistent with CD8+ T cells being considered immune promoting. However, we found that the prognosis was worse in the high-risk group, in which CD8+ T cells were highly expressed. That may be related to its high stromal cell expression. Recent studies have revealed that stromal cell activation in TME is immunosuppressive and that several stromal cell types are considered to have a powerful effect on cancer (33,34). Yu et al. proposed that CAF-driven CD73 is a novel immune checkpoint that reduces the killing effect of CD8+ T cells with decreased IFN-γ production (77). Doedens et al. demonstrated that tumor-associated macrophages impede T-cell activation by enriching the hypoxic TME, leading to an enhanced tumor progression (78). Furthermore, non-classical stem cell-promoting functions of nerves may mediate cancer malignancy in gastrointestinal, pancreatic, and prostate cancers (79-83). Promotion of extracellular matrix and protease secretion by CAFs may facilitate aggressive growth of CC cells (84-88). Thus, we conclude that stromal cell activation is associated with a poorer tumor prognosis. Meanwhile, quite a few researches have implicated that ubiquitination is intimately associated with stromal cell activation. Zhang et al. demonstrated that cisplatin and paclitaxel could promote the secretion of miR-522 from CAFs by activating the USP7/hnRNPA1 axis, leading to the reduction of lipid reactive oxygen species accumulation in cancer cells, and ultimately to the chemotherapy sensitivity decreased (18). That is consistent with our findings that USP7 is a protective gene. Besides, Gao et al. demonstrated that USP7 is essential for endometrial stromal cell metaphase in mice (89). In summary, we hypothesize that stromal cell activation is associated with a poorer prognosis of CC and URGs are involved in this process.
Our study identified and analyzed URGs associated with prognosis in patients with CC. Then, a risk score model related to the prognosis of CC was developed with 7-URG, which showed satisfactory predictive performance. However, there are some limitations in our study. First, the clinical data from GSE17538 are incomplete such as cancer staging and therapy, which might provide clues on biomarker of treatment. Second, specific mechanisms by which URGs affect stromal cell activation in the TME are lacking. The relationship between ubiquitination levels and TME is only a preliminary study, and the specific mechanisms of action and regulatory relationships need further investigation.
In conclusion, we developed and validated prognosis-related prediction models with 7-URG of CC patients for the first time. Higher the risk score of CC patients is, shorter the survival period. We also revealed that tumor ubiquitination was closely associated with stromal cell activation in TME. These results may be enlightening for breaking the bottleneck of CC treatment.
Conclusions
We formed a seven-gene signature prognostic model based on the expression of USP7, UBE2K, USP9X, UBE2B, UBE2E2, USP8, and UBE2G2 in CC, which has excellent predictive power for patient prognosis. Meanwhile, this work revealed the relationship between ubiquitination and stromal cell activation in TME. These results provide a new direction for developing new CC-targeted drugs and optimization of immunotherapy.
Acknowledgments
We acknowledge the TCGA and GEO project for sharing valuable datasets.
Funding: This work was supported by grants from
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-607/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-607/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-607/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lugli A, Zlobec I, Berger MD, et al. Tumour budding in solid cancers. Nat Rev Clin Oncol 2021;18:101-15. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019;16:713-32. [Crossref] [PubMed]
- Wang J, Li S, Liu Y, et al. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer Med 2020;9:361-73. [Crossref] [PubMed]
- Wang Z, Li Y, Mao R, et al. DNAJB8 in small extracellular vesicles promotes Oxaliplatin resistance through TP53/MDR1 pathway in colon cancer. Cell Death Dis 2022;13:151. [Crossref] [PubMed]
- Dong X, Sun Y, Li Y, et al. Synergistic Combination of Bioactive Hydroxyapatite Nanoparticles and the Chemotherapeutic Doxorubicin to Overcome Tumor Multidrug Resistance. Small 2021;17:e2007672. [Crossref] [PubMed]
- Saldana M, VanderVorst K, Berg AL, et al. Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer. Endocr Relat Cancer 2019;26:R1-R14. [Crossref] [PubMed]
- Telerman A, Amson R. The molecular programme of tumour reversion: the steps beyond malignant transformation. Nat Rev Cancer 2009;9:206-16. [Crossref] [PubMed]
- Gross S, Rahal R, Stransky N, et al. Targeting cancer with kinase inhibitors. J Clin Invest 2015;125:1780-9. [Crossref] [PubMed]
- Li X, Yang KB, Chen W, et al. CUL3 (cullin 3)-mediated ubiquitination and degradation of BECN1 (beclin 1) inhibit autophagy and promote tumor progression. Autophagy 2021;17:4323-40. [Crossref] [PubMed]
- Huang Y, Yang X, Lu Y, et al. UBE2O targets Mxi1 for ubiquitination and degradation to promote lung cancer progression and radioresistance. Cell Death Differ 2021;28:671-84. [Crossref] [PubMed]
- Yue M, Yun Z, Li S, et al. NEDD4 triggers FOXA1 ubiquitination and promotes colon cancer progression under microRNA-340-5p suppression and ATF1 upregulation. RNA Biol 2021;18:1981-95. [Crossref] [PubMed]
- Higa LA, Wu M, Ye T, et al. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 2006;8:1277-83. [Crossref] [PubMed]
- Li C, Bu J, Liao Y, et al. High Expressions of CUL4A and TP53 in Colorectal Cancer Predict Poor Survival. Cell Physiol Biochem 2018;51:2829-42. [Crossref] [PubMed]
- Deng L, Meng T, Chen L, et al. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther 2020;5:11. [Crossref] [PubMed]
- Chen F, Zhuang X, Lin L, et al. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med 2015;13:45. [Crossref] [PubMed]
- Guo Y, Yang L, Lei S, et al. NEDD4 Negatively Regulates GITR via Ubiquitination in Immune Microenvironment of Melanoma. Onco Targets Ther 2019;12:10629-37. [Crossref] [PubMed]
- Zhang H, Deng T, Liu R, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer 2020;19:43. [Crossref] [PubMed]
- Feng Y, Zhang Y, Cai Y, et al. A20 targets PFKL and glycolysis to inhibit the progression of hepatocellular carcinoma. Cell Death Dis 2020;11:89. [Crossref] [PubMed]
- O'Malley G, Treacy O, Lynch K, et al. Stromal Cell PD-L1 Inhibits CD8+ T-cell Antitumor Immune Responses and Promotes Colon Cancer. Cancer Immunol Res 2018;6:1426-41. [Crossref] [PubMed]
- Herrero A, Benedicto A, Romayor I, et al. Inhibition of COX-2 Impairs Colon Cancer Liver Metastasis through Reduced Stromal Cell Reaction. Biomol Ther (Seoul) 2021;29:342-51. [Crossref] [PubMed]
- Amara S, Chaar I, Khiari M, et al. Stromal cell derived factor-1 and CXCR4 expression in colorectal cancer promote liver metastasis. Cancer Biomark 2015;15:869-79. [Crossref] [PubMed]
- Shin HN, Moon HH, Ku JL. Stromal cell-derived factor-1α and macrophage migration-inhibitory factor induce metastatic behavior in CXCR4-expressing colon cancer cells. Int J Mol Med 2012;30:1537-43. [Crossref] [PubMed]
- Mustafi R, Dougherty U, Shah H, et al. Both stromal cell and colonocyte epidermal growth factor receptors control HCT116 colon cancer cell growth in tumor xenografts. Carcinogenesis 2012;33:1930-9. [Crossref] [PubMed]
- Xu F, Wang Z, Song X, et al. A Direct and Sensitive Method for Determination of 5-Fluorouracil in Colorectal Cancer Cells: Evaluating the Effect of Stromal Cell on Drug Resistance of Cancer Cells. J Anal Methods Chem 2021;2021:6689488. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015;348:74-80. [Crossref] [PubMed]
- Molon B, Ugel S, Del Pozzo F, et al. Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J Exp Med 2011;208:1949-62. [Crossref] [PubMed]
- Fienberg R. The stromal theca cell and postmenopausal endometrial adenocarcinoma. Cancer 1969;24:32-8. [Crossref] [PubMed]
- Garcia AJ, Ruscetti M, Arenzana TL, et al. Pten null prostate epithelium promotes localized myeloid-derived suppressor cell expansion and immune suppression during tumor initiation and progression. Mol Cell Biol 2014;34:2017-28. [Crossref] [PubMed]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. [Crossref] [PubMed]
- Xie Q, Ding J, Chen Y. Role of CD8+ T lymphocyte cells: Interplay with stromal cells in tumor microenvironment. Acta Pharm Sin B 2021;11:1365-78. [Crossref] [PubMed]
- Elsaleh H, Powell B, Soontrapornchai P, et al. p53 gene mutation, microsatellite instability and adjuvant chemotherapy: impact on survival of 388 patients with Dukes' C colon carcinoma. Oncology 2000;58:52-9. [Crossref] [PubMed]
- Elsaleh H, Joseph D, Grieu F, et al. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet 2000;355:1745-50. [Crossref] [PubMed]
- Hemminki A, Mecklin JP, Järvinen H, et al. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology 2000;119:921-8. [Crossref] [PubMed]
- Jover R, Zapater P, Castells A, et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur J Cancer 2009;45:365-73. [Crossref] [PubMed]
- Carethers JM, Chauhan DP, Fink D, et al. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology 1999;117:123-31. [Crossref] [PubMed]
- Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 2010;138:958-68. [Crossref] [PubMed]
- Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005;23:609-18. [Crossref] [PubMed]
- Suryadinata R, Roesley SN, Yang G, et al. Mechanisms of generating polyubiquitin chains of different topology. Cells 2014;3:674-89. [Crossref] [PubMed]
- Li C, Du L, Ren Y, et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J Exp Clin Cancer Res 2019;38:76. [Crossref] [PubMed]
- Bhattacharya S, Chakraborty D, Basu M, et al. Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases. Signal Transduct Target Ther 2018;3:17. [Crossref] [PubMed]
- Qi SM, Cheng G, Cheng XD, et al. Targeting USP7-Mediated Deubiquitination of MDM2/MDMX-p53 Pathway for Cancer Therapy: Are We There Yet? Front Cell Dev Biol 2020;8:233. [Crossref] [PubMed]
- Rawat R, Starczynowski DT, Ntziachristos P. Nuclear deubiquitination in the spotlight: the multifaceted nature of USP7 biology in disease. Curr Opin Cell Biol 2019;58:85-94. [Crossref] [PubMed]
- Turnbull AP, Ioannidis S, Krajewski WW, et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 2017;550:481-6. [Crossref] [PubMed]
- Kategaya L, Di Lello P, Rougé L, et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 2017;550:534-8. [Crossref] [PubMed]
- Valles GJ, Bezsonova I, Woodgate R, et al. USP7 Is a Master Regulator of Genome Stability. Front Cell Dev Biol 2020;8:717. [Crossref] [PubMed]
- Paudel P, Zhang Q, Leung C, et al. Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X. Proc Natl Acad Sci U S A 2019;116:7288-97. [Crossref] [PubMed]
- Xiong W, Gao X, Zhang T, et al. USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy. Nat Commun 2022;13:1700. [Crossref] [PubMed]
- Dufner A, Kisser A, Niendorf S, et al. The ubiquitin-specific protease USP8 is critical for the development and homeostasis of T cells. Nat Immunol 2015;16:950-60. [Crossref] [PubMed]
- Hormaechea-Agulla D, Kim Y, Song MS, et al. New Insights into the Role of E2s in the Pathogenesis of Diseases: Lessons Learned from UBE2O. Mol Cells 2018;41:168-78. [PubMed]
- Li B, Jiang J, Assaraf YG, et al. Surmounting cancer drug resistance: New insights from the perspective of N6-methyladenosine RNA modification. Drug Resist Updat 2020;53:100720. [Crossref] [PubMed]
- Taketo K, Konno M, Asai A, et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol 2018;52:621-9. [PubMed]
- Somasagara RR, Spencer SM, Tripathi K, et al. RAD6 promotes DNA repair and stem cell signaling in ovarian cancer and is a promising therapeutic target to prevent and treat acquired chemoresistance. Oncogene 2017;36:6680-90. [Crossref] [PubMed]
- Nayar U, Cohen O, Kapstad C, et al. Acquired HER2 mutations in ER+ metastatic breast cancer confer resistance to estrogen receptor-directed therapies. Nat Genet 2019;51:207-16. [Crossref] [PubMed]
- Lyakhovich A, Shekhar MP. Supramolecular complex formation between Rad6 and proteins of the p53 pathway during DNA damage-induced response. Mol Cell Biol 2003;23:2463-75. [Crossref] [PubMed]
- Lyakhovich A, Shekhar MP. RAD6B overexpression confers chemoresistance: RAD6 expression during cell cycle and its redistribution to chromatin during DNA damage-induced response. Oncogene 2004;23:3097-106. [Crossref] [PubMed]
- Shekhar MP, Lyakhovich A, Visscher DW, et al. Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res 2002;62:2115-24. [PubMed]
- Somasagara RR, Tripathi K, Spencer SM, et al. Rad6 upregulation promotes stem cell-like characteristics and platinum resistance in ovarian cancer. Biochem Biophys Res Commun 2016;469:449-55. [Crossref] [PubMed]
- Menezes J, Acquadro F, Wiseman M, et al. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia 2014;28:823-9. [Crossref] [PubMed]
- Pichler A, Knipscheer P, Oberhofer E, et al. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol 2005;12:264-9. [Crossref] [PubMed]
- Donath S, Li P, Willenbockel C, et al. Apoptosis repressor with caspase recruitment domain is required for cardioprotection in response to biomechanical and ischemic stress. Circulation 2006;113:1203-12. [Crossref] [PubMed]
- Foo RS, Chan LK, Kitsis RN, et al. Ubiquitination and degradation of the anti-apoptotic protein ARC by MDM2. J Biol Chem 2007;282:5529-35. [Crossref] [PubMed]
- Bonacci T, Emanuele MJ. Dissenting degradation: Deubiquitinases in cell cycle and cancer. Semin Cancer Biol 2020;67:145-58. [Crossref] [PubMed]
- Yoshida Y, Tanaka K. Cytosolic N-Glycans: Triggers for Ubiquitination Directing Proteasomal and Autophagic Degradation: Molecular Systems for Monitoring Cytosolic N-Glycans as Signals for Unwanted Proteins and Organelles. Bioessays 2018; [Crossref] [PubMed]
- Li S, Zhao J, Shang D, et al. Ubiquitination and deubiquitination emerge as players in idiopathic pulmonary fibrosis pathogenesis and treatment. JCI Insight 2018;3:e120362. [Crossref] [PubMed]
- Kong D, Zhang Z, Chen L, et al. Curcumin blunts epithelial-mesenchymal transition of hepatocytes to alleviate hepatic fibrosis through regulating oxidative stress and autophagy. Redox Biol 2020;36:101600. [Crossref] [PubMed]
- Liao X, Li Y, Liu J, et al. Deubiquitinase USP13 promotes extracellular matrix expression by stabilizing Smad4 in lung fibroblast cells. Transl Res 2020;223:15-24. [Crossref] [PubMed]
- Li Y, Wang C, Xi HM, et al. Chorionic villus-derived mesenchymal stem cells induce E3 ligase TRIM72 expression and regulate cell behaviors through ubiquitination of p53 in trophoblasts. FASEB J 2021;35:e22005. [Crossref] [PubMed]
- Zencheck WD, Xiao H, Weiss LM. Lysine post-translational modifications and the cytoskeleton. Essays Biochem 2012;52:135-45. [Crossref] [PubMed]
- Křivohlavá R, Grobárová V, Neuhöferová E, et al. Interaction of colon cancer cells with glycoconjugates triggers complex changes in gene expression, glucose transporters and cell invasion. Mol Med Rep 2018;17:5508-17. [PubMed]
- He Y, Liu T, Dai S, et al. Tumor-Associated Extracellular Matrix: How to Be a Potential Aide to Anti-tumor Immunotherapy? Front Cell Dev Biol 2021;9:739161. [Crossref] [PubMed]
- Zanotelli MR, Zhang J, Reinhart-King CA. Mechanoresponsive metabolism in cancer cell migration and metastasis. Cell Metab 2021;33:1307-21. [Crossref] [PubMed]
- Chang SC, Zhang BX, Ding JL. E2-E3 ubiquitin enzyme pairing - partnership in provoking or mitigating cancers. Biochim Biophys Acta Rev Cancer 2022;1877:188679. [Crossref] [PubMed]
- Yu M, Guo G, Huang L, et al. CD73 on cancer-associated fibroblasts enhanced by the A2B-mediated feedforward circuit enforces an immune checkpoint. Nat Commun 2020;11:515. [Crossref] [PubMed]
- Doedens AL, Stockmann C, Rubinstein MP, et al. Macrophage expression of hypoxia-inducible factor-1 alpha suppresses T-cell function and promotes tumor progression. Cancer Res 2010;70:7465-75. [Crossref] [PubMed]
- Yamazaki S, Ema H, Karlsson G, et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 2011;147:1146-58. [Crossref] [PubMed]
- Liebig C, Ayala G, Wilks J, et al. Perineural invasion is an independent predictor of outcome in colorectal cancer. J Clin Oncol 2009;27:5131-7. [Crossref] [PubMed]
- Ayala GE, Dai H, Powell M, et al. Cancer-related axonogenesis and neurogenesis in prostate cancer. Clin Cancer Res 2008;14:7593-603. [Crossref] [PubMed]
- Demir IE, Friess H, Ceyhan GO. Nerve-cancer interactions in the stromal biology of pancreatic cancer. Front Physiol 2012;3:97. [Crossref] [PubMed]
- Magnon C, Hall SJ, Lin J, et al. Autonomic nerve development contributes to prostate cancer progression. Science 2013;341:1236361. [Crossref] [PubMed]
- Tommelein J, Verset L, Boterberg T, et al. Cancer-associated fibroblasts connect metastasis-promoting communication in colorectal cancer. Front Oncol 2015;5:63. [Crossref] [PubMed]
- Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature 2004;432:332-7. [Crossref] [PubMed]
- Erez N, Glanz S, Raz Y, et al. Cancer associated fibroblasts express pro-inflammatory factors in human breast and ovarian tumors. Biochem Biophys Res Commun 2013;437:397-402. [Crossref] [PubMed]
- Olumi AF, Grossfeld GD, Hayward SW, et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res 1999;59:5002-11. [PubMed]
- Tsuyada A, Chow A, Wu J, et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res 2012;72:2768-79. [Crossref] [PubMed]
- Gao Y, Wang Y, Zhou C, et al. Ubiquitin-specific protease 7 (USP7) is essential for endometrial stromal cell decidualization in mice. Dev Growth Differ 2019;61:176-85. [Crossref] [PubMed]


