
The prognostic role of fatty acid metabolism-related genes in patients with gastric cancer
Introduction
Gastric cancer (GC) is the fifth most frequent disease in the world and the third major cause of cancer deaths each year, according to statistics (1). Most GC (about 90%) is adenocarcinoma, originating from the stomach’s most superficial or mucosal glands. GC is traditionally classified as intestinal and diffuse histological subtypes (2). The mechanism of GC has not yet been clarified. In 1975, Correa et al. proposed the hypothesis of GC. Many factors may play a role in the aetiology of GC, leading to the transformation of normal stomach tissue from multiple stages to GC. Stimulative environment and other factors, including acid secretion, excessive growth of bacteria, and the production of nitrite or N-nitroso compounds by bacteria from dietary nitrates, jointly lead to the gradual occurrence and metaplasia of normal gastric epithelium and ultimately progress to GC (3,4). Epidemiological studies have shown that family history, diet, drinking, smoking, Helicobacter pylori, and Epstein-Barr virus (EBV) infection are important risk factors for GC (5-8). The incidence of GC is different in gender and geographical differences. The susceptibility of males is two to three times that of females. The incidence of GC shows excellent regional diversity. More than 50% of new GCs occur in developing countries. GC has the highest incidence in Eastern Europe and East Asia (China and Japan) in Central and South America (9). With the deepening research on GC by researchers, significant progress has been made in preventing, examining, and treating GC. Therefore, the incidence of GC in most parts of the world has been declining, and the 5-year survival rate has improved significantly (10-13). Up to now, clinicians still mainly use tumor, lymph node, and metastasis (TNM) staging systems to monitor GC progress, which is also the most widely used prognostic indicator. In addition, some other biomarkers such as mucins (MUC) or human epithelial growth factor receptor-2 (HER2) are also gradually used to predict GC prognosis (14). However, there exists a high degree of heterogeneity between GC, and patients with the same TNM stage or HER2 and MUC expression usually have significant differences in treatment effect, drug resistance, and survival outcome. Therefore, finding novel and reliable factors connected to the survival time of GC patients is an excellent supplement to the previous prognosis prediction system and may be used as an evaluation index of the effect of GC treatment, or even as a target for GC targeted therapy, which has important guiding significance for clinical practice.
In normal human cells, the cytoplasm and mitochondria of cells provide a place for metabolism, resulting in glucose or fatty acids that provide most of the energy for human cells (15). Cancer cells usually have characteristic metabolic changes, and the utilization of glucose and fatty acids will also change accordingly. Previous studies have shown that the Warburg effect in glucose metabolism, especially in glycolysis, plays a vital role in human tumor diseases (16,17). Researchers have gradually noticed the relationship between fatty acid metabolism and cancer in recent years. In mammalian cells, fatty acid can be obtained directly from the surrounding microenvironment and can also be synthesized from nutrients such as glucose or glutamine. In cancer cells, lipid metabolic pathways are extensively reconstructed, including fatty acid transport, ab initio fat formation, storage as lipid droplets (LD), and ATP production by β-oxidation (18). Fatty acid is widely involved in cancer cell energy storage, membrane proliferation, and signal molecule production. Therefore, the metabolic changes of fatty acid can directly affect various behaviors of tumor cells, including proliferation, invasion, and metastasis (19). Based on this, people are committed to studying the related factors regulating fatty acid metabolism to find new anticancer strategies and prognostic factors. However, some scholars have conducted relevant studies on biomarkers related to fatty acid metabolism (20-22). There is, however, a dearth of systematic study that can adequately demonstrate its importance to the prognosis of patients with GC. This study selected the transcriptome profile and clinical survival data of 483 GC samples from different databases. We researched the relationship between fatty acid metabolism-related genes and GC prognosis, comprehensively evaluated the fatty acid metabolism mode, and constructed the risk score model of fatty acid prognosis. Furthermore, the variation in the degree of immune cell infiltration in stomach cancer tissues under different risk scores was investigated in this study. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-761/rc).
Methods
Cohort data of GC patients
This study included two cohorts and 483 samples. Four hundred seven transcriptome data were collected from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) as test sets, including 375 tumor tissue data and 32 non-neoplastic tissue data. The data type is High Throughput Sequencing-fragments per kilobase of transcript per million mapped reads (HTSeq-FPKM). After excluding the data with missing values and survival time less than 30 days, 352 clinical data were obtained. The corresponding clinical data were extracted according to the following contents: age, gender, total staging, grade, TNM staging, survival time, and survival situation.
Seventy-six transcriptome data were collected from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) as validation sets, all of which were GC tissue data. The patients’ survival time and status were extracted as clinical data. The microarray data configuration file was based on platform GSE84426.
Ninety-three common genes related to fatty acid metabolism were identified from the Gene Set Enrichment Analysis (GSEA) database (http://www.gsea-msigdb.org/gsea/index.jsp).
The corresponding genes annotations referred to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) data.
Data processing and validation of the prognostic model
To deal with the fatty acid metabolism-related genes differentially expressed (DEGs) between GC and non-neoplastic tissue in the TCGA cohort, we used the R package “limma”. The retrieved DEGs must match the following criteria: false discovery rate (FDR) of 0.05 and absolute log2 fold change (FC) >0.75. Subsequently, the “ggpubr” package and “corrplot” were used to make the analysis results visualized by boxplots and heatmap.
To investigate the link between fatty acid metabolism-related gene expression and GC prognosis, we divided the patients who came from GC cohort into three different subgroups by using the “Consensus Plus” R package. The survival curve was then generated using the “survival” package based on the Kaplan-Meier analysis to compare the survival outcomes of the subgroups.
Univariate Cox regression was used to determine whether fatty acid metabolism-related genes are connected with overall survival (OS) in patients with GC. The “glmnet” package and “survival” package were used to establish the prognosis model for selected prognostic genes to obtain the association between prognostic genes and patient survival. To select the appropriate prognostic genes for complicated data, the LASSO regression in the “glment” package was employed to minimize the influence of strongly linked genes. Then we included prognostic genes and arrived at the risk score calculation. According to the expression score of prognostic associated genes, patients with GC were separated into two groups: high-risk and low-risk.
Validation of the prognostic model
As previously stated, we classified GC patients into different risk categories based on the expression of prognostic genes. The Kaplan-Meier function was then used to compare the survival outcomes of the two patients and to examine the area under the receiver operating characteristic (ROC) curve (AUC) to ensure the stability of the prognostic model. Following that, we used the TCGA cohort to test the accuracy of the prognostic model in predicting patient outcome to standard prediction approaches, and we used the forest map as a presentation. The clinical factors and prognosis-related genes’ heatmap was plotted to summarize the association between gene expression and clinical practice. The TCGA patients were divided into low-risk and high-risk groups based on the prognostic model score, and differential expression analysis was done. The screened differential genes were enriched and evaluated to see whether there were any variations in gene function between the risk groups. Finally, the difference in the degree of immune cell infiltration in GC tissues between risk groups was examined in order to investigate the association between immune cell infiltration in GC tissues and scores for various risk variables.
Statistical analysis
For all of the aforementioned analysis, a P value of less than 0.05 was considered statistically significant.
Ethical consideration
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Fatty acid metabolism-related genes in GC
All the genes which were including in our study could be acquired in Appendix 1. We observed the gene expression differences in GC patients and non-neoplastic persons in the TCGA cohort and found 30 fatty acid metabolism-related genes. The heatmap clearly revealed the differential expression of the genes selected in patients with GC and non-neoplastic people (Figure 1A). The boxplot allowed for more in-depth examination of the differential expression of genes involved in fatty acid metabolism. The boxplot showed the expression levels of ALDH1B1, ACACB, ADH1B, HACD4, ELOVL4, HACD1, ADH7, ALDH3A2, ACOT2, ACOT1, ADH4, ADH6, ADH1C, and ALDH2 in non-neoplastic people were higher than those in GC patients. The expression of ELOVL7, OXSM, HSD17B12, ACLY, ACACA, FASN, HACD2, HACD3, ACOT7, MCAT, MECR, OLAH, ELOVL2, CPT1C, PPT1, ELOVL3 in non-neoplastic people was lower than that in GC patients (Figure 1B). In addition, we conducted a correlation analysis of these 30 fatty acid metabolism-related genes to observe the potential links between genes.
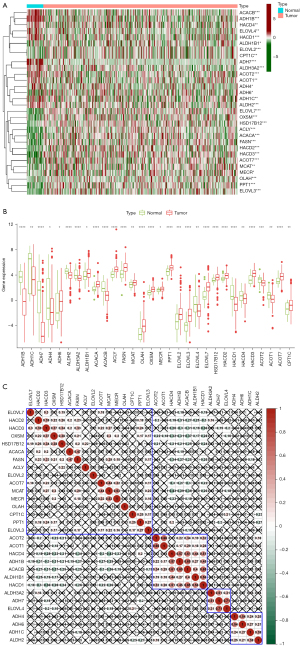
The results of correlation analysis showed that HACD1 had a relation with multiple genes, in which ACACB was the most significant. The expression level of HACD1 was probable to promote the expression of ACACB (Figure 1C).
Use of consensus clustering according to fatty acid metabolism-related genes to identify different clusters of GC patients with different clinical outcomes
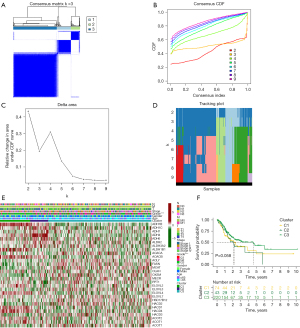
According to the status of fatty acid metabolism-related genes, CG samples were divided into different subgroups to analyze each subgroup’s survival and clinical correlation.
The Consensus Plus analysis in R indicated that when k =3, the difference between clusters is the most minimized (Figure 2A-2D). Therefore, according to the analysis results, we determined the optimal cluster number of 3 and divided patients in TCGA cohorts into different clusters. Subsequently, we made a heat map according to the different clinical data between the three clusters. We analyzed the survival of patients in these three clusters according to the OS of patients. The heatmap showed differences in gene expression between different subgroups (Figure 2E). Kaplan-Meier analysis indicated that the survival time of subgroup 1 and subgroup 2 was similar, which was longer than that of subgroup 3. However, we did not find the difference statistically significant (Figure 2F).
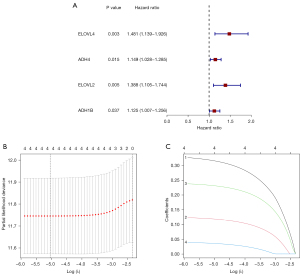
Generation of LASSO model
In order to research the relevance of fatty acid metabolism-related genes to the GC prognosis, we used univariate and LASSO Cox regression to select four best prognosis related genes from 30 fatty acid metabolism-related genes: ELOVL4 (coefficient 0.31), CPT1C (coefficient 0.12), ADH4 (coefficient 0.13), ADH1B (coefficient 0.04) (Figure 3A-3C). Therefore, we obtained the risk scoring formula: risk score = (0.31 × ELOVL4 expression value) + (0.12 × CPT1C expression value) + (0.13 × ADH4 expression value) + (0.04 × ADH1B expression value).
Verifying the reliability of the prognosis model
To investigate the prognostic impact of these four prognostic genes, we split GC patients into distinct risk groups based on prognostic model scores. Following that, we preliminary validated the prognosis model. The Kaplan-Meier survival analysis was repeated based on the risk score groupings. The Kaplan-Meier survival analysis revealed that, regardless of whether the cohort was TCGA or GEO, the OS of the high-risk group was much lower than that of the low-risk group, and the results were statistically significant (Figure 4A,4B). In addition, in order to facilitate the observation of the impact of a single gene on the OS of patients, we again conducted a survival analysis of a single gene in patients with TCGA cohort. Survival analysis showed that each prognosis-related gene could well predict the prognosis of patients, which further proved the accuracy of our prognosis model (Figure 4C-4F).
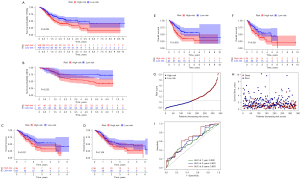
Then we made the risk score distribution map of GC patients, each point representing the survival outcome of each patient (Figure 4G,4H). In addition, we employed the ROC curve to assess the accuracy of our prognostic model’s prediction in TCGA patients. The results indicated that AUC of 1-, 3-, and 5-year OS was more significant than 0.5 (0.590; 0.622; 0.663) (Figure 4I), indicating that these four prognostic-related genes as biomarkers of GC have the solid predictive ability for the prognosis of GC.
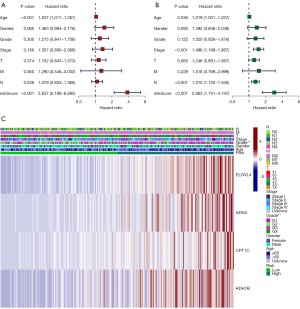
Prognostic model and clinicopathological features
After excluding cases with missing clinical pathology data and those with a survival time of less than 30 days from the cohort, Cox regression analysis was done. Univariate analysis revealed that, among the standard methods for evaluating the prognosis of GC patients, only stage was connected with the prognosis of GC patients, and the risk score had a greater predictive effect than stage (Figure 5A). Then, under the influence of many clinical parameters, we performed a multivariate Cox regression analysis to see if the prognostic model retained its reliable predictive abilities. The findings of the multivariate Cox regression analysis verified the risk score’s dependability once more (Figure 5B), indicating that the risk score may be utilised as an effective supplement to standard prediction methods to more correctly forecast the prognosis of GC patients. Therefore, we believed that our prognostic model could not only accurately evaluate the prognosis of GC as an independent prognostic means but also provide more valuable judgment for the prognosis of patients with traditional prediction means. Finally, we made a clinical correlation heatmap according to the prognostic model and clinicopathological features. The heatmap showed that the expression of prognostic genes was much lower in the low-risk group than in the high-risk group (Figure 5C), confirming the accuracy of our prognosis model.
Function of DEGs in different risk-groups
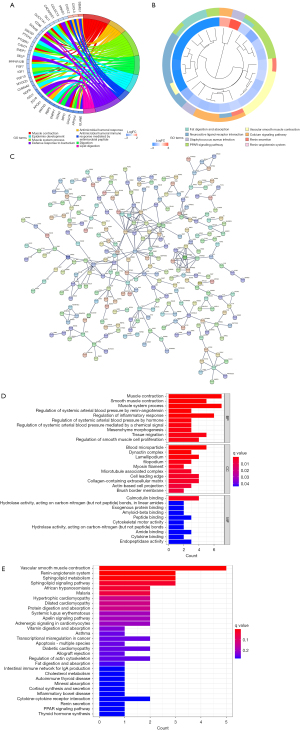
We split patients in the TCGA cohort into high-risk and low-risk groups and evaluated the difference in gene expression between the two groups to detect DEGs (P<0.05, logFC >0.75). In the high-risk and low-risk groups, 884 genes were found to be differently expressed. GO and KEGG enrichment analysis for displaying primary functions of DEGs. GO enrichment analysis indicated that gene functions were mainly expressed in the muscle system, immune defense, and fatty acid metabolism (Figure 6A). KEGG enrichment analysis indicated that the gene function was mainly manifested in fat metabolism and digestion, receptor-ligand pathway, and smooth muscle contraction (Figure 6B). In addition, we performed protein interaction network analysis on all differential genes in String (Figure 6C) and extracted the first 20 genes with the strongest association for enrichment analysis again. GO and KEGG enrichment analysis demonstrated that gene functions were mainly enriched in smooth muscle contraction, sphingomyelin metabolism, and inflammatory response regulation (Figure 6D,6E), proving that high and low-risk groups were grouped was indeed associated with fatty acid metabolism.
Fatty acid metabolism-related genes and immune cell
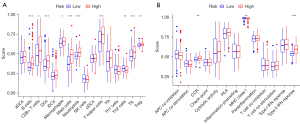
Given that the enrichment analysis results indicated that the role of immune cells in patients with varying risk scores may differ, we examined tissue immune cell infiltration in patients in low and high-risk groups. The results showed that the infiltration levels of B cells, Dendritic cells, T helper cells, mast cells, neutrophils, and tumor-infiltrating lymphocytes were much lower in the low-risk group than in the high-risk group (Figure 7A). Immune cells in the high-risk group had a more robust type II IFN response, CCR, and MHC I receptor function than those in the low-risk group, and the difference was statistically significant (Figure 7B). This shows that immune cells may influence the prognosis of GC patients with fatty acid metabolism-related genes.
Discussion
People have achieved more advances in basic research and clinical diagnosis and treatment of stomach cancer thanks to the tireless efforts of researchers, but the diagnosis, treatment, and prognosis of GC patients remain unsatisfactory (23-25). Therefore, it is necessary to clarify the exact molecular mechanism of GC to find possible therapeutic targets and prognostic markers for GC. More and more researchers focus on the role of fatty acid metabolism (including synthesis and catabolic reactions) in cancer. Many researches have demonstrated that abnormal expression of genes related to fatty acid synthesis or oxidation is associated with malignant phenotypes, including metastasis, treatment resistance, and recurrence (26). Researchers found that the energy preference of cancer cells for glycolysis even in aerobic conditions and the hypoxic environment caused by the imbalance of vascular distribution by interstitial tumor fibrosis often cause acidosis in tumors (27-29), acidosis promotes or inhibits acetylation of mitochondria and histones in cancer cells, thus altering fatty acid metabolism and promoting cancer cell proliferation (30). Due to the lack of previous studies on GC and fatty acid metabolism-related genes, it is not easy to effectively prove the relationship between them at this stage. Therefore, we screened DEGs in GC patients in this study and then selected four genes related to prognosis to establish a prognosis model. The prognosis model was extensively validated to produce a score model capable of reliably predicting the prognosis of GC patients and providing innovative ideas for future GC research.
According to our study, four genes were included in the establishment of the prognosis model. Overexpression of these four genes indicated a poor prognosis, indicating that these four genes might play an essential part during different processes of GC progression. The full name of ELOVL4 is elongation of very-long-chain fatty acids-4. Its encoding product is a long chain enzyme that is responsible for the production of ultra-long chain saturated fatty acids and polyunsaturated fatty acids in the brain, retina, skin, meibomian gland, and testis (31). ELOVL4 mutations have been linked to significant system disorders such as Stargardt’s macular dystrophy (STGD3) and spinocerebellar ataxia 34 (SCA34) (32). In addition, ELOVL4 may be involved in maintaining of the skin permeability barrier and sperm function (33). In addition to neuroblastoma (34), we did not find the relationship between ELOVL4 and other tumors. Therefore, according to our findings, we support further studies on the relationship between ELOVL4 and different tumors such as GC to reveal the possible mechanism of ELOVL4 in GC progression.
Carnitine palmitoyltransferase 1C (CPT1C) regulates mitochondrial energy metabolism as well as tumor cell growth. Chen et al. (35) confirmed that CPT1C promotes GC progression by mediating enhanced fatty acid oxidation. CPT1C was also found to play a role in GC resistance (36). Previously, it was shown that a new miR-1291-ERR-CPT1C axis plays a role in cancers such as breast cancer and pancreatic cancer, suggesting a new carcinogenic mechanism of CPT1C (37). ADH4 and ADH1B are different subtypes of alcohol dehydrogenase, related to many physiological and pathological processes in humans, including alcohol metabolism, liver function damage, and cancer occurrence (38). ADH4 and ADH1B are associated with various gastrointestinal tumors, especially hepatocellular carcinoma, colorectal cancer, GC, and esophageal cancer (39-42). However, the existing studies have not clarified the molecular association between ADH4 and ADH1B and the pathogenesis of GC.
Our study provides an accurate prediction model in addition to the traditional forecasting methods for the prognosis of GC patients. We have carried out a variety of verifications to ensure its scientificity. In addition, previous studies have shown that up-regulated or down-regulated genes in GC can induce the infiltration of immune cells (43-45). We investigated immune cell infiltration in GC tissues under various risk variables. We discovered that up-regulation of prognosis-related genes may promote the infiltration of B cells, dendritic cells, auxiliary T cells, mast cells, neutrophils, and tumor-infiltrating lymphocytes in GC tissues, which provided a novel idea for further research into immune cell infiltration in GC tissues.
The limitations of this study are mainly manifested in the fact that all the analyses are carried out using the sample data of TCGA and GEO, and no experiments are carried out to prove our research conclusions. In addition, the large sample size gap between tumor tissue and non-neoplastic tissue in the TCGA database may lead to potential errors in differential gene screening results. In addition, this study fails to cover all countries and races and cannot avoid the existence of selective bias. Therefore, it is still uncertain whether there is universality in all countries and races.
Conclusions
In summary, we demonstrated that the increase of ELOVL4, CPT1C, ADH4, and ADH1B gene levels was closely related to OS prolongation in GC patients. Furthermore, using univariate and LASSO Cox regression analysis, we developed a prognostic model involving ELOVL4, CPT1C, ADH4, and ADH1B gene expression and demonstrated its stability and repeatability. We also believe that these four genes play a major role in GC prognosis prediction and can be exploited as therapeutic targets. We believe that the conclusions of our research can provide more insights for more and more prospective GC treatment literature.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-761/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-761/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Birkman EM, Mansuri N, Kurki S, et al. Gastric cancer: immunohistochemical classification of molecular subtypes and their association with clinicopathological characteristics. Virchows Arch 2018;472:369-82. [Crossref] [PubMed]
- Correa P, Haenszel W, Cuello C, et al. A model for gastric cancer epidemiology. Lancet 1975;2:58-60. [Crossref] [PubMed]
- Correa P, Cuello C, Duque E, et al. Gastric cancer in Colombia. III. Natural history of precursor lesions. J Natl Cancer Inst 1976;57:1027-35. [Crossref] [PubMed]
- Yang K, Lu L, Liu H, et al. A comprehensive update on early gastric cancer: defining terms, etiology, and alarming risk factors. Expert Rev Gastroenterol Hepatol 2021;15:255-73. [Crossref] [PubMed]
- Guggenheim DE, Shah MA. Gastric cancer epidemiology and risk factors. J Surg Oncol 2013;107:230-6. [Crossref] [PubMed]
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700-13. [Crossref] [PubMed]
- Venerito M, Link A, Rokkas T, et al. Gastric cancer - clinical and epidemiological aspects. Helicobacter 2016;21:39-44. [Crossref] [PubMed]
- Machlowska J, Baj J, Sitarz M, et al. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci 2020;21:4012. [Crossref] [PubMed]
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700-13. [Crossref] [PubMed]
- Zhao Q, Cao L, Guan L, et al. Immunotherapy for gastric cancer: dilemmas and prospect. Brief Funct Genomics 2019;18:107-12. [Crossref] [PubMed]
- Wu D, Zhang P, Ma J, et al. Serum biomarker panels for the diagnosis of gastric cancer. Cancer Med 2019;8:1576-83. [Crossref] [PubMed]
- Sterea AM, Egom EE, El Hiani Y. TRP channels in gastric cancer: New hopes and clinical perspectives. Cell Calcium 2019;82:102053. [Crossref] [PubMed]
- Machlowska J, Maciejewski R, Sitarz R. The Pattern of Signatures in Gastric Cancer Prognosis. Int J Mol Sci 2018;19:1658. [Crossref] [PubMed]
- Judge A, Dodd MS. Metabolism. Essays Biochem 2020;64:607-47. [Crossref] [PubMed]
- Abbaszadeh Z, Çeşmeli S, Biray Avcı Ç. Crucial players in glycolysis: Cancer progress. Gene 2020;726:144158. [Crossref] [PubMed]
- Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 2013;12:152. [Crossref] [PubMed]
- Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer 2020;122:4-22. [Crossref] [PubMed]
- Currie E, Schulze A, Zechner R, et al. Cellular fatty acid metabolism and cancer. Cell Metab 2013;18:153-61. [Crossref] [PubMed]
- Jiang M, Wu N, Xu B, et al. Fatty acid-induced CD36 expression via O-GlcNAcylation drives gastric cancer metastasis. Theranostics 2019;9:5359-73. [Crossref] [PubMed]
- Lee JY, Nam M, Son HY, et al. Polyunsaturated fatty acid biosynthesis pathway determines ferroptosis sensitivity in gastric cancer. Proc Natl Acad Sci U S A 2020;117:32433-42. [Crossref] [PubMed]
- Tan Y, Lin K, Zhao Y, et al. Adipocytes fuel gastric cancer omental metastasis via PITPNC1-mediated fatty acid metabolic reprogramming. Theranostics 2018;8:5452-68. [Crossref] [PubMed]
- Pasechnikov V, Chukov S, Fedorov E, et al. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol 2014;20:13842-62. [Crossref] [PubMed]
- Zeng Z, Yang B, Liao Z. Progress and prospects of immune checkpoint inhibitors in advanced gastric cancer. Future Oncol 2021;17:1553-69. [Crossref] [PubMed]
- Mohri Y, Toiyama Y, Kusunoki M. Progress and prospects for the discovery of biomarkers for gastric cancer: a focus on proteomics. Expert Rev Proteomics 2016;13:1131-9. [Crossref] [PubMed]
- Kuo CY, Ann DK. When fats commit crimes: fatty acid metabolism, cancer stemness and therapeutic resistance. Cancer Commun (Lond) 2018;38:47. [Crossref] [PubMed]
- Ji K, Mayernik L, Moin K, et al. Acidosis and proteolysis in the tumor microenvironment. Cancer Metastasis Rev 2019;38:103-12. [Crossref] [PubMed]
- Pillai SR, Damaghi M, Marunaka Y, et al. Causes, consequences, and therapy of tumors acidosis. Cancer Metastasis Rev 2019;38:205-22. [Crossref] [PubMed]
- Damgaci S, Ibrahim-Hashim A, Enriquez-Navas PM, et al. Hypoxia and acidosis: immune suppressors and therapeutic targets. Immunology 2018;154:354-62. [Crossref] [PubMed]
- Corbet C, Pinto A, Martherus R, et al. Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab 2016;24:311-23. [Crossref] [PubMed]
- Hopiavuori BR, Anderson RE, Agbaga MP. ELOVL4: Very long-chain fatty acids serve an eclectic role in mammalian health and function. Prog Retin Eye Res 2019;69:137-58. [Crossref] [PubMed]
- Deák F, Anderson RE, Fessler JL, et al. Novel Cellular Functions of Very Long Chain-Fatty Acids: Insight From ELOVL4 Mutations. Front Cell Neurosci 2019;13:428. [Crossref] [PubMed]
- Yeboah GK, Lobanova ES, Brush RS, et al. Very long chain fatty acid-containing lipids: a decade of novel insights from the study of ELOVL4. J Lipid Res 2021;62:100030. [Crossref] [PubMed]
- Rugolo F, Bazan NG, Calandria J, et al. The expression of ELOVL4, repressed by MYCN, defines neuroblastoma patients with good outcome. Oncogene 2021;40:5741-51. [Crossref] [PubMed]
- Chen T, Wu G, Hu H, et al. Enhanced fatty acid oxidation mediated by CPT1C promotes gastric cancer progression. J Gastrointest Oncol 2020;11:695-707. [Crossref] [PubMed]
- Maeda O, Ando T, Ohmiya N, et al. Alteration of gene expression and DNA methylation in drug-resistant gastric cancer. Oncol Rep 2014;31:1883-90. [Crossref] [PubMed]
- Chen Y, Zhou Y, Han F, et al. A novel miR-1291-ERRα-CPT1C axis modulates tumor cell proliferation, metabolism and tumorigenesis. Theranostics 2020;10:7193-210. [Crossref] [PubMed]
- Polimanti R, Gelernter J. ADH1B: From alcoholism, natural selection, and cancer to the human phenome. Am J Med Genet B Neuropsychiatr Genet 2018;177:113-25. [Crossref] [PubMed]
- Seol JE, Kim J, Lee BH, et al. Folate, alcohol, ADH1B and ALDH2 and colorectal cancer risk. Public Health Nutr 2020; Epub ahead of print. [Crossref] [PubMed]
- Ishioka K, Masaoka H, Ito H, et al. Association between ALDH2 and ADH1B polymorphisms, alcohol drinking and gastric cancer: a replication and mediation analysis. Gastric Cancer 2018;21:936-45. [Crossref] [PubMed]
- Wei RR, Zhang MY, Rao HL, et al. Identification of ADH4 as a novel and potential prognostic marker in hepatocellular carcinoma. Med Oncol 2012;29:2737-43. [Crossref] [PubMed]
- Yang SJ, Yokoyama A, Yokoyama T, et al. Relationship between genetic polymorphisms of ALDH2 and ADH1B and esophageal cancer risk: a meta-analysis. World J Gastroenterol 2010;16:4210-20. [Crossref] [PubMed]
- Zhang AZ, Yuan X, Liang WH, et al. Immune Infiltration in Gastric Cancer Microenvironment and Its Clinical Significance. Front Cell Dev Biol 2022;9:762029. [Crossref] [PubMed]
- Bai X, Wong CC, Pan Y, et al. Loss of YTHDF1 in gastric tumors restores sensitivity to antitumor immunity by recruiting mature dendritic cells. J Immunother Cancer 2022;10:e003663. [Crossref] [PubMed]
- Zhou X, Fang D, Liu H, et al. PMN-MDSCs accumulation induced by CXCL1 promotes CD8+ T cells exhaustion in gastric cancer. Cancer Lett 2022;532:215598. [Crossref] [PubMed]


