
Utilization of model-agnostic explainable artificial intelligence frameworks in oncology: a narrative review
Introduction
In oncology, commonly used statistical models include linear regression, logistic regression, and Cox-proportional hazards regression. These produce odds ratios, hazard ratios, coefficients, and P values, which are relatively to interpret and apply. Oncology is an increasingly data driven field with nuanced clinical questions, which in some cases necessitates more complicated models, such as modelling non-linear and/or distribution assumptions-free relationships, interaction effects, or image analysis, to better utilize data and make more informed and accurate decisions. As processing power has increased, machine learning (ML) algorithms have the potential to produce improved and clinically valuable models (1). However, as models increase in complexity relative to common regressions models, they become increasingly difficult for end-users (inclusive of clinicians, patients, administrators, stakeholders, and more) to understand, turning more into a metaphorical “black box” (2). End-users do not just care that the output of a model is accurate; they also want to know how that output is produced and might be influenced (3). Therein lies a primary difficulty of using ML algorithms for treatment decisions in the oncology clinic, limiting an otherwise powerful tool (4). The problem is three-fold: interpretability of the models, transparency of the ML algorithms, and sensitivity of the algorithms/models to the minute changes in the data and algorithmic parameters (5).
This dilemma of models losing interpretability as complexity increases has led to so called “explainable artificial intelligence (XAI)” techniques, which broadly seek to improve understanding of complex models by using interpretable visualizations and sets of rules to represent the inner workings of the ML “black box” and pinpoint the most salient and consequential features of the models (Figure 1) (6-8). The way XAI works differs based on the type of data it is explaining. In structured data analysis, XAI aims to identify the variables (and their interactions) that influenced the model output. In image analysis, XAI aims to identify the regions of interest which influenced the model output. Commonly used XAI frameworks include Local Interpretable Model-agnostic Explanations (LIME) (9) and SHapley Additive exPlanations (SHAP) (10), which have found success in fields such as finance (11), insurance (12), and healthcare and, despite their differences in implementation, both aim to improve interpretability of ML models (13). Details of SHAP and LIME can be found in Table 1. Of note, several other XAI frameworks do exist, including class activation mapping (CAM) or gradient weighted (Grad)CAM frameworks, but these are not model-agnostic, meaning they can only be applied to specific ML models (convolution neural networks in the case of CAM and GradCAM) (14). At their core, these frameworks aim to quantify the contribution that each feature brings to the prediction made by a model. In theory, they can work with any type of ML model to better understand how features yield given predictions.

Table 1
| Framework | Overview | Pros | Cons |
|---|---|---|---|
| SHAP | Based on Shapley values from game theory | Can single prediction explanations or global model interpretations | Explanations of non-tree-based algorithms can be computationally expensive |
| Calculates the average marginal contribution of a feature value over all possible combinations of predictions and sets of inputs | Explanations of tree-based algorithms are computationally inexpensive and exact | ||
| Perturbs data globally to build a model that is accurate locally (analogous to Fisher’s exact test) | Can guarantee consistency and local accuracy | ||
| Bottom line: decompose the final prediction into the contribution of each attribute | Diverse array of available summary plots | ||
| LIME | Based on assumption that models behave linearly on a local scale | Computationally efficient for most algorithms | Less exhaustive so cannot guarantee accuracy and consistency |
| Builds sparse linear models around an individual prediction in its local vicinity of inputs | Useful for explaining individual predictions | Plots primarily limited to single prediction | |
| Perturbs data around an individual prediction (locally) to build a model that is accurate locally | Explanations are approximations | ||
| Functionally a subset of SHAP (analogous to chi-squared test) |
Code does not natively support some ML algorithms such as XGBoost | ||
| Bottom line: tells you, in a local sense, what is the most important attribute around the data point of interest |
SHAP, SHapley Additive exPlanations; LIME, Local Interpretable Model-agnostic Explanations; ML, machine learning.
In addition to healthcare in general, ML models and XAI have been combined and applied to clinical oncologic questions. However, use of XAI in oncology is still in an early stage and not widely recognized, and possible applications have yet to be summarized. Therefore, we reviewed the use of XAI in oncology literature, which will optimally provide a framework for possible future applications. In the interest of limiting scope, we limited results to SHAP and LIME, which are the primary model-agnostic XAI frameworks that are more generally applicable. We present the following article in accordance with the Narrative Review reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1626/rc).
Methods
We performed a non-systematic review of the latest literature to characterize utilization of XAI in oncology research. We included relevant articles in English available in the MEDLINE/PubMed database up to 01 May 2022. Search terms included (“explainable artificial intelligence” OR “XAI” OR “EAI” OR “SHAP” OR “LIME”) AND (“oncology” OR “cancer”). In this article, we summarize use cases in oncology wherein XAI has been published to aid in ML model interpretability, which can translate to improved adoption in the clinic (Table 2).
Table 2
| Items | Specification |
|---|---|
| Date of search | 01 May 2022 |
| Databases and other sources searched | MEDLINE/PubMed |
| Search terms used | (“explainable artificial intelligence” OR “XAI” OR “EAI” OR “SHAP” OR “LIME”) AND (“oncology” OR “cancer”) |
| Timeframe | Beginning of database through 01 May 2022 |
| Inclusion and exclusion criteria | All study types and reviews, written in English language |
| Selection process | Consensus between co-author |
XAI, explainable artificial intelligence; EAI, explainable artificial intelligence; SHAP, SHapley Additive exPlanations; LIME, Local Interpretable Model-agnostic Explanations.
Utilization of XAI in oncology research
Prognostication
Perhaps the area of oncology that has been most extensively explored using XAI is prognostication, which is of no surprise given the abundance of large data sets [including the National Cancer Database (NCDB) and the Surveillance, Epidemiology, and End Results (SEER)] with outcomes data. These are of great interest to both clinicians and patients, given that this approach not only can help counsel patients on prognosis and planning, but might also inform on potential interventions that can lead to improvements in outcomes.
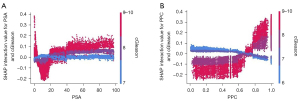
A clinical example that lends itself nicely to the utility of XAI is modeling of interactions between disease characteristics and prostate cancer and their impact on survival. Li et al. modeled the impact of prostate specific antigen (PSA), percent positive cores (PPCs), and Gleason score on survival using the extreme gradient boosted (XGB) tree algorithm (15). Specifically, they sought to examine nonlinear relationships and interactions, which facilitates identification of prognostic thresholds. Since LIME primarily looks only at individual predictions, SHAP tends to be the main framework to perform such analyses. Although the impact of the specified factors on survival was not controversial, modeling with XAI revealed nuances that contradict modern risk stratification. Visualization of such interactions is only possible by using a more complicated model than standard regressions and then explaining that model. For example, when examining the interaction between percentage of positive cores and Gleason score, the SHAP dependence plots revealed that PPC is largely irrelevant if Gleason score is 7 or less. Additionally, when Gleason score is 8 or higher, a PPC threshold of 0.7 best illustrates differences in outcomes, as opposed to the threshold of 0.5 that is used in modern practice to differentiate between favorable and unfavorable intermediate risk prostate cancer. The plots are also able to illustrate phenomena such as patients with high Gleason score and exceedingly low PSA counterintuitively having inferior survival compared to patients with more intermediate PSA levels. These example plots can be found in Figure 2. In these interaction plots, the combined effect of the two variables are plotted, with a value of zero representing an overall neutral effect.
Bertsimas et al. also used XAI to visualize nonlinear relationships, thereby identifying prognostic thresholds. The authors used an XGB model and the SHAP framework to explore the impact of lymph node ratio on survival compared to lymph node count in pancreatic cancer, which is used in the current American Joint Committee on Cancer (AJCC) staging system (16). Their model using lymph node ratio outperformed the AJCC schema at predicting 1-year overall survival with an area under the curve (AUC) of 0.638 vs. 0.586 (though not validated via DeLong’s test or confidence intervals). Using the SHAP dependence plots, the authors, via inspection, identified thresholds for lymph node ratio and tumor size. The authors’ identification of relevant lymph node ratio thresholds was feasible by using graphical interpretation of dependence plots generated by the SHAP framework, and given the interaction is nonlinear, identification of thresholds with linear methods such as logistic regression or Cox regression would have required some trial and error and would not have been as precise.
The aforementioned studies primarily examined XAI plots that summarize all predictions within the datasets. XAI also has been used to examine prognostication in individual predictions (i.e., patients). Both SHAP and LIME have this functionality implemented. In a study by Jansen et al., both SHAP and LIME are used to explain an XGB model of 10-year overall survival in breast cancer patients (17). In this study, the authors use LIME to model individual patients and predictions of overall survival, which explains how individual patients’ characteristics yield their prediction for 10-year overall survival. They did the same with SHAP, but as detailed in Table 1, SHAP has improved functionality in illustrating global impact of features in all patients, so they also presented a summary plot. Lastly, they compared all individual patient explanations produced by LIME and SHAP, demonstrating agreement in 87.8% to 99.9% (95.4% overall) of cases depending on the feature examined. This study highlights key distinctions between LIME and SHAP; both approaches can yield consistent local results, but SHAP is able to examine global trends within models. The same approach, examining individual patients and overall trends has been performed on models predicting survival with nasopharyngeal cancer tumor burden (18).
Lastly, perhaps the way XAI was used most commonly for prognostication and oncology in general was to delineate global feature importance by summarizing all model predictions and outputting each feature’s mean impact across all predictions. This permits a general understanding of how the overall model functions, similar to the summary statistics generated by regression models. Moncada-Torres et al. used XAI to explain a general model of overall survival in breast cancer (19). Importantly, models such as Cox regression are routinely used for such applications, with outputs being readily interpretable by most end-users. ML algorithms, namely the XGB tree algorithm significantly outperformed Cox regression. However, the standard output of such an algorithm is not as readily understandable in terms of how its output is computed. In this example, the authors used the SHAP framework to identify feature importance within the model, which can be compared to intuition to build trust in the model. By using the SHAP framework, the study was able to illustrate the prognostic significance of multiple commonly used variables, thereby facilitating adoption of algorithms such as XGB.
Several other studies have used similar approaches to the aforementioned study to create powerful prognostic models to illustrate nuanced interactions that influence prognosis, with XAI used to explain their ML model on a global level. Additional examples include predicting 30-day mortality following colorectal cancer surgery (20), 5-year survival and esophageal cancer (21), characterizing influence of ethnicity on outcomes and multiple myeloma (22), predicting hospital length of stay (23), and risk of skeletal related events following discontinuation of denosumab among patients with bone metastases (24).
Diagnosis
An additional area of research interest is ways to improve diagnosis of cancer, where ML models have proven to be valuable tools, but given the high stakes of a cancer diagnosis, it is important that the predictions of such models are both accurate and easy to explain to clinicians and patients, representing a great opportunity for XAI.
In one such application, Suh et al. explored a model predicting prostate cancer in general as well as clinically significant prostate cancer prior to prostate biopsy (25). In this study, the authors used XAI frameworks for one of the reasons detailed in the previous section (understanding global feature importance), as well as a new one (feature selection/construction for building clinically-relevant tools). When explained using SHAP summary visualizations, the authors identified important features predicting prostate cancer and clinically significant prostate cancer, which included known predictive factors such as PSA and Gleason score, and how changing these factors individually influenced risk. These visualizations facilitated translation to a risk calculator, via aiding selection of salient features, that could be deployed as a data driven risk estimator (https://boramae-pcrc.appspot.com/) that would be generally applicable to the oncology clinic.
In another study, Kwong et al. reported on a model that predicted side-specific extraprostatic extension and pre-prostatectomy patients (26), again using SHAP summaries to gauge global feature importance. Furthermore, the authors examined the non-linear relationships between relevant factors and probability of site-specific extraprostatic extension with dependence plots, which is of clinical relevance given that features such as PPCs were relatively noncontributory until reaching approximately 75% based on inspection of dependence plots. Though these conclusions are qualitative in nature, they inform quantitative hypotheses that can inform other statistical approaches and overall clinical intuition. Thus, the XAI complemented the ML model by not only providing information to surgeons on risk of extraprostatic extension but also providing explanations of how that risk was calculated for given patient, where identification of non-linear interactions is valuable.
Radiomics
A primary field of oncology that has benefited greatly from XAI is radiomics, given that it has the reputation of being a “black box” or “fishing expedition” (27,28). Radiomics is defined as a process designed to extract quantitative features from imaging, which can subsequently be used for hypothesis generation, testing, or both (29). In oncology, this might mean using imaging characteristics to predict tumor molecular features, behavior, and prognosis. In doing so, a patient’s image might be input into a radiomic algorithm and predict risk of relapse. However accurate such a prediction might be, it is also of vital importance that providers be able to understand why the imaging produces such a prediction.
Radiomics specifically represents a field where LIME might be preferred in certain scenarios, given that radiomics is highly interested in individual predictions and commonly uses non-tree-based algorithms, meaning SHAP becomes computationally intensive. Several radiomics studies focus on using XAI and focusing on individual predictions. One example that has been explored in the literature is molecular classification of gliomas. In gliomas, molecular subtypes related to isocitrate dehydrogenase (IDH) mutations and 1p/19q co-deletion are of vital relevance to prognosis and might also inform treatment options (30). Although confirmation of such abnormalities occurs via pathologic analysis, it can be useful to identify patients with such abnormalities prior to surgery or if pathologic conformation is not possible. Manikis et al. reported on radiomic patterns that can predict IDH mutational status with an accuracy of 73.6% and an associated sensitivity and specificity of 0.6 and 0.736, respectively (31). Following model development, results were explained using both the SHAP and LIME frameworks, which were able to identify important radiomic features that lead to an image being classified as IDH-mutant, which the authors were then able to correlate with biological behavior of IDH-mutant gliomas.
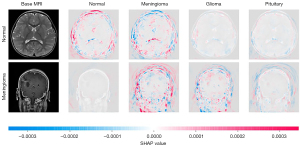
XAI can also be used to describe how a given imaging voxel contributes to a model’s output. Gaur et al. used a deep learning model to identify brain tumor subtypes, with accuracy of 94.64% (32). To explain how their model made its predictions, they provide examples using the SHAP framework where SHAP values are superimposed on imaging voxels, and in doing so illustrate graphically and intuitively how example images are classified as normal or meningioma, as illustrated in Figure 3. In these images, the probability of a given classification is increased with red pixels and decreased with blue pixels. As can be seen, for the normal MRI, the normal classification has the greatest amount of red pixels. The same idea holds true for the meningioma classification. The authors use both SHAP and LIME, again because this classification problem in highly interested in individual cases. Another similar published application is classification of ultrasound imaging of lymph nodes to predict nodal metastasis and early breast cancer (achieving an accuracy of 81.05%, which used LIME to graphically identify regions of interest in individual images) (33).
In addition to explaining the radiomic features, XAI also can aid in development of the models. A major problem in ML is overfitting training data, which can be a result of feeding the excessive features in the algorithm, including features that are largely irrelevant or strongly correlated with the causative features. This is particularly relevant in radiomics, where massive amounts of data points might be generated from a given image set. A challenge XAI helps address is identifying only the most important features predictive of the outcome, which improves final model feature selection, thus eliminating the irrelevant ones, leading to a better model that is more robust and less prone to overfitting. Kha et al. have reported on identification of a radiomic signature predictive of 1p/19q co-deletion (34). During model development, the SHAP framework was used to identify the most important features to include in their model, leading to improved model performance when less important features, based on average SHAP values, were removed, with an AUC of 0.710 before feature selection and 0.753 after.
Other applications of combination ML and XAI in radiomics simply use the resulting visualizations to identify global feature importance, to better understand and build trust in their respective models. These include prediction of early progression of nasopharyngeal cancer following intensity-modulated radiation therapy using MRI (which used the SHAP framework to demonstrate that select radiomic features were more predictively powerful than staging) (35), classification of breast cancer molecular subtypes using non-contract computed tomography (achieving an accuracy of 71.3%, using SHAP to identify important radiomic features) (36). In total, these explanations of radiomic models allows providers to better understand what the model is looking at, which eases application to the clinic by presenting them as more than abstract “black boxes”.
Pathology
Pathology is another diagnostic specialty in oncology where XAI has improved predictive ML models. Like radiomics, pathology is also a field highly interested in individual predictions, where both SHAP and LIME can often be applicable. In a study similar to the one performed by Gaur et al. (32) in radiomics, Palatnik de Sousa et al. used XAI to depict how a neural network identified tumors in histology samples of lymph node metastases (37). Given that this problem involved a convolution neural network and was primarily interested in individual patients, this was a prime example where LIME was a good choice, where SHAP would be more computationally inefficient. Using this method, the model output can overlay histology slides, and end-users can see locations of interest identified by the model. These overlays permitted the authors to biologically validate results by comparing highlighted areas of interest with clinical intuition. This technique has also been applied to diagnosing leukemia (accuracy of 98.38%) (38). Examination of individual patients has also been used on structured data to help classify primary and metastatic cancers using origin-based DNA methylation profiles (39).
Next, pathology studies have harnessed plots of non-linear interactions to identify prognostic thresholds or make a case for policy change. Chakraborty et al. examined the influence of the tumor microenvironment on prognosis in breast cancer (40). In this study, not only did XAI facilitate identification of prognostic factors via global feature importance, but it also illustrated non-linear interactions that permitted identification prognostic thresholds for each factor, which could help tailor identification of treatments that could manipulate the tumor microenvironment and improve prognosis. In this study, the authors used SHAP dependence plots to identify inflection points to correspond to potentially clinically relevant thresholds, generally corresponding to where SHAP values crossed zero. Similarly, in a paper examining the impact of synoptic reporting on survival in patients with prostate cancer using an XGB model explained by SHAP, Janssen et al. reported (41). Where this study has the possibility to benefit pathology is using XAI summary plots, the authors could conclude that synoptic reporting, specifically reporting pathologic data in a structured manner, is the second most important factor after age. In this example, these plots allow interpretation of a ML model that could lead to real policy changes, such as standard implementation of synoptic reporting, that would be difficult to illustrate to decision makers without XAI.
Lastly, as is the case in other disciplines, XAI has been used to explain overall models to better understand how they generally function. Meena et al. developed a model for diagnosis of squamous cell carcinoma (SCC) based on genetic signature (42). In a simple example of how SHAP values can be powerful, when distinguishing between healthy tissue and SCC, only a single gene (HNRNPM) produced a significant impact on the model, which was interestingly not among the 20 most important genes in actinic keratosis based on SHAP values. Although accurate diagnosis of SCC is useful, and the model was 92.86% accurate, these signatures made available by XAI are highly valuable to end-users evaluating reports without access to sophisticated models, permitting powerful qualitative conclusions. Similar approaches have been applied to hematopoietic cancer subtype classification (accuracy of 97.01%) (43).
Treatment selection
Following diagnosis, an area where XAI has a significant opportunity to influence the clinic is in treatment selection. This might include identification of optimal treatment options and predicting outcomes of a given intervention. This is of great interest to the oncology clinic, as any additional information to determine optimal patient care is appreciated, but understanding such information is equally important in order to be able to explain recommendations to patients, their family, and other members of the care team.
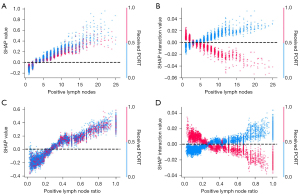
Dependence plots generated by XAI can be useful for identification of predictive thresholds. Ladbury et al. (44) and Zarinshenas et al. (45) examined the prognostic and predictive value of nodal burden in endometrial cancer and locally advanced non-small cell lung cancer (NSCLC), respectively, with associated XAI plots aiding in addressing controversies in the field. In endometrial cancer, via qualitative inspection of SHAP plots, XAI facilitated identification of a threshold of four or more positive nodes where treatment with adjuvant chemoradiation achieved optimal outcomes, while chemotherapy alone had a neutral effect and radiation alone had a deleterious effect. This finding adds insight following publication of PORTEC-3 and GOG 258, wherein optimal adjuvant therapy and sequencing remains unclear (46,47). In locally advanced NSCLC, again via qualitative inspection of SHAP plots, XAI enabled identification of nodal thresholds including three or more positive lymph nodes or a lymph node ration of 0.34 or greater as possible scenarios where addition of postoperative radiotherapy might improve outcomes, which is an area of controversy following publication of the LungART and PORT-C trials suggesting no benefit with postoperative radiotherapy (48,49). These conclusions were only possible because XAI enabled graphical depiction of interactions between nodal burden and treatments in the models, allowing for identification of predictive thresholds. The associated plots illustrating these interactions are found in Figure 4. Using a value of zero as neutral, the aforementioned thresholds can be identified, where patients who do not receive postoperative radiotherapy, represented in blue, have increased risk above those thresholds.
Beyond predicting optimal treatments, XAI has also demonstrated ability to help globally explain models that predict outcomes of treatments. Namely, Laios et al. (50) and Bang et al. (51) explored models that predicted for complete cytoreduction in ovarian cancer and curative resection in early undifferentiated gastric cancer. In the case of ovarian cancer, the model predicted R0 resection with an AUC of 0.866, using SHAP to identify important features. Additionally, using SHAP it identified non-linear interactions via inspection of dependence plots, such as significant decreases in R0 resection rates with peritoneal carcinomatosis indices greater than five and no significant change in R0 resection rates in years from 2017 onwards. In the case of gastric cancer, the model predicted curative resection with accuracy of 89.8% and used SHAP to identify important factors. In combination, these two studies using XAI provide end-users with useful information that can help counsel patients not only on odds of curative resection but also explain how these estimates were determined.
Epidemiology
An additional area involves improvements in epidemiology analyses using XAI, which benefit from examining global interactions and individual predictions. Ahmed et al. used ML to explore spatial variability of lung and bronchus cancer mortality rates across the contiguous United States (52). The authors used dependence plots, break down plots, and maps to visually and geographically represent how key factors were interrelated and might affect specific geographic locations, rather than simply describing the United States population as a whole. For example, they use XAI not only to show that Union County, Florida has an almost 13-fold higher risk of mortality than Summit County, Utah, but also show that elevation is the largest protective factor in Summit County, while smoking is the largest risk factor in Union County. These conclusions were made possible by the authors using waterfall plots for corresponding individual explanations to visualize how model behavior can vary drastically based on the given example. This information permits the model to be both predictive and able to inform possible interventions. In a study by Kobylińska et al., XAI was used to investigate the influence of factors on lung cancer screening (53). The authors used the SHAP framework to produce summary plots, to overall illustrate lung cancer risk in lung cancer screening populations, dependence plots to show how changing individual variables influenced risk, and plots of individual predictions, which can be used how individual patient risk is calculated. This information is useful, as it can be broadly used to counsel patients on decreasing lung cancer risk, but can also inform specific patients of the best way to decrease their risk by identifying the most impactful factors to them.
Similar to other disciplines, studies in epidemiology have also benefitted from XAI aiding with feature selection/construction. In a study by Richter et al., electronic health record (EHR) data was utilized to predict risk of developing melanoma (54). In the study, use of XAI not only depicted important factors that are associated with risk of developing melanoma, but it also facilitated improvements in model efficiency that would facilitate use of ML algorithms on EHR data by helping to select only the most relevant and impactful features, which would otherwise require prohibitive computational resources due to large data size and data missingness.
Radiation treatment workflow
One more niche area where XAI has been evaluated is in the radiation oncology clinic aiding in workflow for treatment planning. The first scenario that was evaluated by Siciarz et al. was clinical decision support systems for radiation plan evaluation in brain tumors (55). The authors used XAI to look at the model globally, to examine interactions within features, and evaluate individual predictions. This model classified treatment plans based on whether the target volume planning objective was met or whether the target volume planning objective was met or not due to a priority trade off due to organ at risk constraints. Such a model uses knowledge from previous radiation treatment plans to inform on when trade-offs might be necessary. SHAP was used on the model to be able to determine relevant dosimetric factors that would inform whether a plan was acceptable, which can provide useful feedback to medical physicist and radiation oncologists when determining ways to further optimize plans that are not deemed acceptable. In this case, XAI was helpful in that it provides information on generally what leads to objectives being met, and also providing a breakdown of specific cases. Next, once the plan is approved, quality assurance is required to ensure patient’s safety and avoid clinically significant errors such as delivery of the desired dose. This is a process that can be somewhat automated with ML models. Again, it is important that a model predicts that a plan can be safely delivered and for end-users to understand why this conclusion was made to facilitate improvements. Chen et al. explored a model that evaluated whether a plan would be deemed acceptable and used XAI to identify features that led to such a prediction, both globally and individually, which aids in further automating the quality assurance process (56).
Discussion
The studies discussed above, including how model-agnostic XAI was used by the authors, is summarized in Table 3. We found that the use of XAI could be divided into five categories: delineation of global feature importance, characterization of individual prediction feature importance, visualization of nonlinear relationships and interactions, identification of prognostic and/or predictive thresholds, and feature selection/construction. These permit additional conclusions to be drawn from ML models, which are key to implementation in the oncology clinic, given that they improve understandability and therefore confidence.
Table 3
| Study | Cancer type | XAI Framework | Purpose of XAI | ||||
|---|---|---|---|---|---|---|---|
| Delineation of global feature importance | Characterization of individual prediction feature importance | Visualization of nonlinear relationships and interactions | Identification of prognostic and/or predictive thresholds | Feature selection | |||
| Prognostication | |||||||
| Li et al., 2020 (15) | Prostate | SHAP | √ | √ | |||
| Bertsimas et al., 2021 (16) | Pancreatic | SHAP | √ | √ | |||
| Jansen et al., 2020 (17) | Breast | SHAP, LIME | √ | √ | √ | ||
| Chen et al., 2021 (18) | Nasopharyngeal | SHAP | √ | √ | √ | ||
| Moncada-Torres et al., 2021 (19) | Breast | SHAP | √ | ||||
| van den Bosch et al., 2021 (20) | Colorectal | SHAP | √ | ||||
| Gong et al., 2021 (21) | Esophageal | SHAP | √ | √ | |||
| Farswan et al., 2021 (22) | Multiple myeloma | SHAP | √ | √ | |||
| Alsinglawi et al., 2022 (23) | Lung | SHAP | √ | ||||
| Jacobson et al., 2022 (24) | Bone metastases | SHAP | √ | √ | |||
| Diagnosis | |||||||
| Suh et al., 2020 (25) | Prostate | SHAP | √ | √ | |||
| Kwong et al., 2022 (26) | Prostate | SHAP | √ | √ | |||
| Radiomics | |||||||
| Manikis et al., 2021 (31) | Glioma | SHAP, LIME | √ | √ | |||
| Gaur et al., 2022 (32) | Brain tumor | SHAP, LIME | √ | ||||
| Lee et al., 2021 (33) | Breast | LIME | √ | ||||
| Kha et al., 2021 (34) | Glioma | SHAP | √ | √ | |||
| Du et al., 2019 (35) | Nasopharyngeal | SHAP | √ | √ | |||
| Wang et al., 2022 (36) | Breast | SHAP | √ | ||||
| Pathology | |||||||
| Palatnik de Sousa et al., 2019 (37) | General | LIME | √ | ||||
| Abir et al., 2022 (38) | Leukemia | LIME | √ | ||||
| Modhukar et al., 2021 (39) | Breast | LIME | √ | ||||
| Chakraborty et al., 2021 (40) | Breast | SHAP, LIME | √ | √ | √ | ||
| Janssen et al., 2022 (41) | Prostate | SHAP | √ | √ | |||
| Meena et al., 2022 (42) | Skin | SHAP | √ | ||||
| Park et al., 2021 (43) | Leukemia | SHAP | √ | ||||
| Treatment selection | |||||||
| Ladbury et al., 2022 (44) | Endometrial | SHAP | √ | √ | |||
| Zarinshenas et al., 2022 (45) | Lung | SHAP | √ | √ | |||
| Laios et al., 2022 (50) | Ovarian | SHAP | √ | √ | √ | ||
| Bang et al., 2021 (51) | Gastric | SHAP | √ | ||||
| Epidemiology | |||||||
| Ahmed et al., 2021 (52) | Lung | SHAP | √ | √ | |||
| Kobylińska et al., 2022 (53) | Lung | SHAP | √ | √ | √ | ||
| Richter et al., 2019 (54) | Melanoma | SHAP | √ | √ | |||
| Radiation treatment workflow | |||||||
| Siciarz et al., 2021 (55) | Brain tumor | SHAP | √ | √ | √ | ||
| Chen et al., 2022 (56) | Prostate | SHAP, LIME | √ | √ | |||
XAI, explainable artificial intelligence; SHAP, SHapley Additive exPlanations; LIME, Local Interpretable Model-agnostic Explanations.
In general, XAI is a vibrant area of ML research, with hundreds of papers produced in the field over the last decade. While there is a wide adoption of interpretability “add-ons” (to the “classical” ML classifiers and estimators), such as SHAP and LIME, there is yet no unifying XAI framework in the broad ML field, let alone biomedical ML. This situation is likely to change, given the heightened interest in XAI within both theoretical and applied ML research communities. Adoption of the cutting-edge XAI ML research advances and methods, preferably biomedical data-specific, in oncology is the next frontier. That being said, there are already existing methods and algorithms in the ML/AI toolkit that are more explainable and interpretable by design, such as probabilistic causal modeling, various Bayesian methodologies and fast/oblique decision trees. Adaptation of such techniques to the oncology spaces is an emerging trend (57-61). Notably, Bayesian network modeling aims to construct and visualize graphical probabilistic/causal multiscale models from the “flat” multivariate data; while outside of the scope of this communication, there is significant recent body of work in the Bayesian networks in oncology space, numbering ~100 publications per annum in the 2020s.
Other areas of active research with direct ties to oncology are computation of hazard ratios from XAI output (62) and using XAI as an explicit feature selection/construction mechanism (63). XAI offers an opportunity for collaboration of oncologists and computer scientists to improve patient care. Integration of EHRs can facilitate training and implementation of models and associated explanations to leverage information and decision-making for oncology patients. With availability of massive quantities of patient data in the EHR, inclusive of genomic and clinical characteristics, applications to better understanding precision medicine that arises from out ML models.
In summary, there remains a fundamental tradeoff between the predictive accuracy (oftentimes directly correlated with the model complexity and opaqueness) and descriptive interpretability (64). While the focus of the ML research has often been on the former, the latter is no less significant in our (oncology) context. Finding the proper balance between the two has been elusive; recent advances in XAI, while promising, remain incremental and fragmentary. Therefore, our broad recommendation to the oncology researchers and practitioners is three-fold: first, utilize the high-performance scalable ML methods augmented with the interpretability tools, such as SHAP and LIME. Second, adopt inherently high-interpretability methods, such as “simple” decision trees, regression models such as least absolute shrinkage and selection operator (LASSO), and probabilistic causal networks, when the data modalities and dimensionality allow such applications and their predictive performance is not that different from more complex (and opaque) models. Third, be on a lookout for the emerging XAI advances in the ML spaces, both systemic and method-specific. By using these approaches, the power of ML can be optimally utilized to improve the oncology clinic. These will help harness ongoing advances in oncology, including big data and personalized medicine, to be able to provide better, personalized, more economic, and less-demanding/toxic diagnostic and treatment options.
Although XAI is a powerful tool for improving the interpretability and thereby adoption of ML models in the oncology clinic, it is not without limitations, which do also apply to oncology research (65). First, as discussed previously, an active area of research is implementation of models that are both accurate and interpretable, which potentially bypassed the need for XAI entirely. Therefore, the notion that ML inherently is a black box that must be explained must not always be true. Nevertheless, less interpretable models continue to be popular in oncology, as detailed by this review, meaning XAI still has a niche to fill as long as the gap between using accurate and interpretable models is being bridged. Next, there are inherent inaccuracies in explanations of certain ML models; many “explanations” are actually approximations or probabilistic measures, as creating exact representations is computationally prohibitive (9,10). Furthermore, attempts to distill models may necessitate simplifications in order to be interpretable, which can raise concerns for applicability as well as human error when the explanations are used to make modifications for individual cases. Of course, this is a major hindrance to adoption in a high-stakes field such as oncology. When addressing this problem, a critical question for researchers is model selection; “simpler” models such as LASSO regression may yield superior generalization results (thanks to decreased overfitting) relative to feature selection facilitated with XAI and a complex ML model. Notably, XAI complements feature selection (or includes feature selection, in the explainability-embedded ML methods), not “competes” with it. A potential analysis framework might include variable selection, then XAI, then further variable selection guided by XAI if necessary. Future work should also seek to characterize “significance” of resulting model explanations, which might be accomplished with a permutation test such as a permutation LASSO for variable selection (66) or aforementioned efforts to generate hazard ratios and p-values from explanations (62). Lastly, the explanations can only be as good as their models, so it is important that models undergo robust testing with suitable performance metrics and are empirically sound before being explained, yielding improved efficacy and safety. Notably, a major limitation of most of the included studies in this review is a lack external validation of model results as well as cross-validation of explanations. Future work should ensure models are extensively quality controlled, tested, and validated, optimally via a schema such as transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) (67), minimum information about clinical artificial intelligence modeling (MI-CLAIM) (68), or radiomics quality score (69), of course acknowledging that due to availability of suitable datasets opportunities for external validation may be limited. Despite these limitations, it is not our recommendation that XAI have no place in oncology. On the contrary, as discussed above it is our belief that it is a valuable tool, but it does need to be implemented responsibly and assessed critically before being used to influence patient care.
Conclusions
ML certainly offers many opportunities for the oncology clinic to be improved. In addition to providing accurate predictive models, it is also important that models be interpretable by providers who will be using them, or else adoption in the clinic will be limited due to their complicated and often indecipherable nature. XAI methodology such as LIME and SHAP can produce powerful and diverse visualizations to illustrate the inner workings of ML algorithms in several oncologic fields, which makes them easier for average end-users to understand, and in some cases provides actionable information that end-users might use to improve patient outcomes. Further, XAI facilitates feature selection/construction, identification of prognostic and/or predictive thresholds, and overall confidence in the models, among other benefits. To ensure ML oncologic research achieves its maximal benefit and reach, future studies should consider utilization of XAI frameworks, which can make the models more understandable to end-users without technical acumen that would otherwise be needed to interpret ML literature.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1626/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1626/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1626/coif). AA serves as an unpaid editorial board member of Translational Cancer Research from December 2019 to November 2023. CL reports grant funding from RefleXion Medical. ASR reports funding from NIH NLM grant R01LM013138, NIH NLM grant R01LM013876, NIH NCI grant U01CA232216 and support from Dr. Susumu Ohno Endowed Chair in Theoretical Biology. AA has grant funding from AstraZeneca. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med 2019;380:1347-58. [Crossref] [PubMed]
- Papernot N, McDaniel P, Goodfellow I, et al. editors. Practical black-box attacks against machine learning. In: Proceedings of the 2017 ACM on Asia Conference on Computer and Communications Security, 2017.
- Diprose WK, Buist N, Hua N, et al. Physician understanding, explainability, and trust in a hypothetical machine learning risk calculator. J Am Med Inform Assoc 2020;27:592-600. [Crossref] [PubMed]
- Price WN. Big data and black-box medical algorithms. Sci Transl Med 2018;10:eaao5333. [Crossref] [PubMed]
- Vilone G, Longo L. Explainable artificial intelligence: a systematic review. arXiv preprint 2020. arXiv:200600093.
- Gunning D, Stefik M, Choi J, et al. XAI-Explainable artificial intelligence. Sci Robot 2019;4:eaay7120. [Crossref] [PubMed]
- Holzinger A, Biemann C, Pattichis CS, et al. What do we need to build explainable AI systems for the medical domain? arXiv preprint 2017. arXiv:171209923.
- Doshi-Velez F, Kim B. Towards a rigorous science of interpretable machine learning. arXiv preprint 2017. arXiv:170208608.
- Ribeiro MT, Singh S, Guestrin C. editors. “Why should I trust you?” Explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016.
- Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Advances in Neural Information Processing Systems, 2017.
- Mokhtari KE, Higdon BP, Başar A. editors. Interpreting financial time series with SHAP values. In: Proceedings of the 29th Annual International Conference on Computer Science and Software Engineering, 2019.
- Kuo K, Lupton D. Towards explainability of machine learning models in insurance pricing. arXiv preprint 2020. arXiv:200310674.
- Lundberg SM, Nair B, Vavilala MS, et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng 2018;2:749-60. [Crossref] [PubMed]
- van der Velden BHM, Kuijf HJ, Gilhuijs KGA, et al. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med Image Anal 2022;79:102470. [Crossref] [PubMed]
- Li R, Shinde A, Liu A, et al. Machine Learning-Based Interpretation and Visualization of Nonlinear Interactions in Prostate Cancer Survival. JCO Clin Cancer Inform 2020;4:637-46. [Crossref] [PubMed]
- Bertsimas D, Margonis GA, Huang Y, et al. Toward an Optimized Staging System for Pancreatic Ductal Adenocarcinoma: A Clinically Interpretable, Artificial Intelligence-Based Model. JCO Clin Cancer Inform 2021;5:1220-31. [Crossref] [PubMed]
- Jansen T, Geleijnse G, Van Maaren M, et al. Machine Learning Explainability in Breast Cancer Survival. Stud Health Technol Inform 2020;270:307-11. [PubMed]
- Chen X, Li Y, Li X, et al. An interpretable machine learning prognostic system for locoregionally advanced nasopharyngeal carcinoma based on tumor burden features. Oral Oncol 2021;118:105335. [Crossref] [PubMed]
- Moncada-Torres A, van Maaren MC, Hendriks MP, et al. Explainable machine learning can outperform Cox regression predictions and provide insights in breast cancer survival. Sci Rep 2021;11:6968. [Crossref] [PubMed]
- van den Bosch T, Warps AK, de Nerée Tot Babberich MPM, et al. Predictors of 30-Day Mortality Among Dutch Patients Undergoing Colorectal Cancer Surgery, 2011-2016. JAMA Netw Open 2021;4:e217737. [Crossref] [PubMed]
- Gong X, Zheng B, Xu G, et al. Application of machine learning approaches to predict the 5-year survival status of patients with esophageal cancer. J Thorac Dis 2021;13:6240-51. [Crossref] [PubMed]
- Farswan A, Gupta A, Sriram K, et al. Does Ethnicity Matter in Multiple Myeloma Risk Prediction in the Era of Genomics and Novel Agents? Evidence From Real-World Data. Front Oncol 2021;11:720932. [Crossref] [PubMed]
- Alsinglawi B, Alshari O, Alorjani M, et al. An explainable machine learning framework for lung cancer hospital length of stay prediction. Sci Rep 2022;12:607. [Crossref] [PubMed]
- Jacobson D, Cadieux B, Higano CS, et al. Risk factors associated with skeletal-related events following discontinuation of denosumab treatment among patients with bone metastases from solid tumors: A real-world machine learning approach. J Bone Oncol 2022;34:100423. [Crossref] [PubMed]
- Suh J, Yoo S, Park J, et al. Development and validation of an explainable artificial intelligence-based decision-supporting tool for prostate biopsy. BJU Int 2020;126:694-703. [Crossref] [PubMed]
- Kwong JCC, Khondker A, Tran C, et al. Explainable artificial intelligence to predict the risk of side-specific extraprostatic extension in pre-prostatectomy patients. Can Urol Assoc J 2022;16:213-21. [Crossref] [PubMed]
- Incoronato M, Aiello M, Infante T, et al. Radiogenomic Analysis of Oncological Data: A Technical Survey. Int J Mol Sci 2017;18:805. [Crossref] [PubMed]
- Liu Z, Wang S, Dong D, et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics 2019;9:1303-22. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol 2017;134:505-12. [Crossref] [PubMed]
- Manikis GC, Ioannidis GS, Siakallis L, et al. Multicenter DSC-MRI-Based Radiomics Predict IDH Mutation in Gliomas. Cancers (Basel) 2021;13:3965. [Crossref] [PubMed]
- Gaur L, Bhandari M, Razdan T, et al. Explanation-Driven Deep Learning Model for Prediction of Brain Tumour Status Using MRI Image Data. Front Genet 2022;13:822666. [Crossref] [PubMed]
- Lee YW, Huang CS, Shih CC, et al. Axillary lymph node metastasis status prediction of early-stage breast cancer using convolutional neural networks. Comput Biol Med 2021;130:104206. [Crossref] [PubMed]
- Kha QH, Le VH, Hung TNK, et al. Development and Validation of an Efficient MRI Radiomics Signature for Improving the Predictive Performance of 1p/19q Co-Deletion in Lower-Grade Gliomas. Cancers (Basel) 2021;13:5398. [Crossref] [PubMed]
- Du R, Lee VH, Yuan H, et al. Radiomics Model to Predict Early Progression of Nonmetastatic Nasopharyngeal Carcinoma after Intensity Modulation Radiation Therapy: A Multicenter Study. Radiol Artif Intell 2019;1:e180075. [Crossref] [PubMed]
- Wang F, Wang D, Xu Y, et al. Potential of the Non-Contrast-Enhanced Chest CT Radiomics to Distinguish Molecular Subtypes of Breast Cancer: A Retrospective Study. Front Oncol 2022;12:848726. [Crossref] [PubMed]
- Palatnik de Sousa I, Maria Bernardes Rebuzzi Vellasco M, Costa da Silva E. Local Interpretable Model-Agnostic Explanations for Classification of Lymph Node Metastases. Sensors (Basel) 2019;19:2969. [Crossref] [PubMed]
- Abir WH, Uddin MF, Khanam FR, et al. Explainable AI in Diagnosing and Anticipating Leukemia Using Transfer Learning Method. Comput Intell Neurosci 2022;2022:5140148. [Crossref] [PubMed]
- Modhukur V, Sharma S, Mondal M, et al. Machine Learning Approaches to Classify Primary and Metastatic Cancers Using Tissue of Origin-Based DNA Methylation Profiles. Cancers (Basel) 2021;13:3768. [Crossref] [PubMed]
- Chakraborty D, Ivan C, Amero P, et al. Explainable Artificial Intelligence Reveals Novel Insight into Tumor Microenvironment Conditions Linked with Better Prognosis in Patients with Breast Cancer. Cancers (Basel) 2021;13:3450. [Crossref] [PubMed]
- Janssen FM, Aben KK, Heesterman BL, et al. Using Explainable Machine Learning to Explore the Impact of Synoptic Reporting on Prostate Cancer. Algorithms 2022;15:49. [Crossref]
- Meena J, Hasija Y. Application of explainable artificial intelligence in the identification of Squamous Cell Carcinoma biomarkers. Comput Biol Med 2022;146:105505. [Crossref] [PubMed]
- Park KH, Batbaatar E, Piao Y, et al. Deep Learning Feature Extraction Approach for Hematopoietic Cancer Subtype Classification. Int J Environ Res Public Health 2021;18:2197. [Crossref] [PubMed]
- Ladbury C, Li R, Shiao J, et al. Characterizing impact of positive lymph node number in endometrial cancer using machine-learning: A better prognostic indicator than FIGO staging? Gynecol Oncol 2022;164:39-45. [Crossref] [PubMed]
- Zarinshenas R, Ladbury C, McGee H, et al. Machine learning to refine prognostic and predictive nodal burden thresholds for post-operative radiotherapy in completely resected stage III-N2 non-small cell lung cancer. Radiother Oncol 2022;173:10-8. [Crossref] [PubMed]
- Matei D, Filiaci V, Randall ME, et al. Adjuvant Chemotherapy plus Radiation for Locally Advanced Endometrial Cancer. N Engl J Med 2019;380:2317-26. [Crossref] [PubMed]
- de Boer SM, Powell ME, Mileshkin L, et al. Adjuvant chemoradiotherapy versus radiotherapy alone for women with high-risk endometrial cancer (PORTEC-3): final results of an international, open-label, multicentre, randomised, phase 3 trial. Lancet Oncol 2018;19:295-309. [Crossref] [PubMed]
- Le Pechoux C, Pourel N, Barlesi F, et al. Postoperative radiotherapy versus no postoperative radiotherapy in patients with completely resected non-small-cell lung cancer and proven mediastinal N2 involvement (Lung ART): an open-label, randomised, phase 3 trial. Lancet Oncol 2022;23:104-14. [Crossref] [PubMed]
- Hui Z, Men Y, Hu C, et al. Effect of Postoperative Radiotherapy for Patients With pIIIA-N2 Non-Small Cell Lung Cancer After Complete Resection and Adjuvant Chemotherapy: The Phase 3 PORT-C Randomized Clinical Trial. JAMA Oncol 2021;7:1178-85. [Crossref] [PubMed]
- Laios A, Kalampokis E, Johnson R, et al. Explainable Artificial Intelligence for Prediction of Complete Surgical Cytoreduction in Advanced-Stage Epithelial Ovarian Cancer. J Pers Med 2022;12:607. [Crossref] [PubMed]
- Bang CS, Ahn JY, Kim JH, et al. Establishing Machine Learning Models to Predict Curative Resection in Early Gastric Cancer with Undifferentiated Histology: Development and Usability Study. J Med Internet Res 2021;23:e25053. [Crossref] [PubMed]
- Ahmed ZU, Sun K, Shelly M, et al. Explainable artificial intelligence (XAI) for exploring spatial variability of lung and bronchus cancer (LBC) mortality rates in the contiguous USA. Sci Rep 2021;11:24090. [Crossref] [PubMed]
- Kobylińska K, Orłowski T, Adamek M, et al. Explainable Machine Learning for Lung Cancer Screening Models. Applied Sciences 2022;12:1926. [Crossref]
- Richter AN, Khoshgoftaar TM. Efficient learning from big data for cancer risk modeling: A case study with melanoma. Comput Biol Med 2019;110:29-39. [Crossref] [PubMed]
- Siciarz P, Alfaifi S, Uytven EV, et al. Machine learning for dose-volume histogram based clinical decision-making support system in radiation therapy plans for brain tumors. Clin Transl Radiat Oncol 2021;31:50-7. [Crossref] [PubMed]
- Chen Y, Aleman DM, Purdie TG, et al. Understanding machine learning classifier decisions in automated radiotherapy quality assurance. Phys Med Biol 2022; [Crossref] [PubMed]
- Kalet AM, Doctor JN, Gennari JH, et al. Developing Bayesian networks from a dependency-layered ontology: A proof-of-concept in radiation oncology. Med Phys 2017;44:4350-9. [Crossref] [PubMed]
- Trilla-Fuertes L, Gámez-Pozo A, Arevalillo JM, et al. Bayesian networks established functional differences between breast cancer subtypes. PLoS One 2020;15:e0234752. [Crossref] [PubMed]
- Park SB, Hwang KT, Chung CK, et al. Causal Bayesian gene networks associated with bone, brain and lung metastasis of breast cancer. Clin Exp Metastasis 2020;37:657-74. [Crossref] [PubMed]
- Wang X, Branciamore S, Gogoshin G, et al. New Analysis Framework Incorporating Mixed Mutual Information and Scalable Bayesian Networks for Multimodal High Dimensional Genomic and Epigenomic Cancer Data. Front Genet 2020;11:648. [Crossref] [PubMed]
- Djulbegovic B, Hozo I, Dale W. Transforming clinical practice guidelines and clinical pathways into fast-and-frugal decision trees to improve clinical care strategies. J Eval Clin Pract 2018;24:1247-54. [Crossref] [PubMed]
- Sundrani S, Lu J. Computing the Hazard Ratios Associated With Explanatory Variables Using Machine Learning Models of Survival Data. JCO Clin Cancer Inform 2021;5:364-78. [Crossref] [PubMed]
- Marcílio WE, Eler DM. editors. From explanations to feature selection: assessing SHAP values as feature selection mechanism. In: 2020 33rd SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), 2020.
- Duval A. Explainable artificial intelligence (XAI). 2019. Available online: https://www.researchgate.net/profile/Alexandre-Duval-2/publication/332209054_Explainable_Artificial_Intelligence_XAI/links/5ca6269aa6fdcca26dfec0cd/Explainable-Artificial-Intelligence-XAI.pdf
- Rudin C. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat Mach Intell 2019;1:206-15. [Crossref] [PubMed]
- Yang S, Wen J, Eckert ST, et al. Prioritizing genetic variants in GWAS with lasso using permutation-assisted tuning. Bioinformatics 2020;36:3811-7. [Crossref] [PubMed]
- Moons KG, Altman DG, Reitsma JB, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1-73. [Crossref] [PubMed]
- Norgeot B, Quer G, Beaulieu-Jones BK, et al. Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat Med 2020;26:1320-4. [Crossref] [PubMed]
- Sanduleanu S, Woodruff HC, de Jong EEC, et al. Tracking tumor biology with radiomics: A systematic review utilizing a radiomics quality score. Radiother Oncol 2018;127:349-60. [Crossref] [PubMed]


