The mechanism study of Eag1 potassium channel in gastric cancer
Introduction
Gastric cancer (GC) is a key reason for cancer morbidity and mortality worldwide, accounting for more than 1080,000 incidences every year and an estimated 768,000 deaths in 2020 (1,2). Although treatment and diagnosis of GC have been improved recently, the 5-year overall survival (OS) rate is still relatively low (3). As a result, GC has become a major contributor to the burden of cancer, accounting for 20% worldwide, following lung and liver cancer (4). As a kind of heterogeneous disease, GC regulation can be affected by heredity and complex molecular mechanisms (5). Several reports have shown that heredity can affect cancer progression, accounting for 10% of GC cases (6,7). Therefore, exploring the specific molecular markers for biological mechanism exploration of GC is important and urgent.
The abnormal regulation of key genes in potassium channels has been found in several kinds of cancer. Among these potassium channels, voltage-gated ether à-go-go-1 (Eag1) has received more attention for tumor pathogenesis in recent years because of its close relation with cell proliferation, survival, angiogenesis, migration, and invasion of cancer cells (8-10). Interestingly, higher expression of Eag1 has been found in most human tumors than in normal tissues (11,12). Further, abnormal expression of Eag1 has been reported to result in a strong immune response in breast cancer (13,14). Moreover, many biological studies have provided strong evidence of the importance of Eag1 in tumor development (15,16). Hence, this attractive feature of restricted expression in normal tissues suggests that Eag1 may be a useful individualized marker in tumor diagnosis, mechanism research and targeted drug therapy. To date, few studies have reported the effect of Eag1 expression on the oncogenic potential in GC.
The objective of this study was to explore and validate the influence and the role of Eag1 channel on the susceptibility to GC. The relationship between Eag1 expression and cell biology was analyzed in GC cells. Biological experiments were performed to estimate the influence of Eag1 knockdown on cell function. Bioinformatics analysis was analyzed for Eag1 validation with multiple public databases. We also further evaluated the influence of different Eag1 expression levels on cyclin D1 as well as cyclin E to explore GC progression. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2276/rc).
Methods
Cell culture
The GC cell lines SGC-7901 and BGC-823 were provided and identified by American Type Culture Collection (ATCC; Manassas, VA, USA). Cells with a passage time less than 6 months were used in this study. Fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) was used to culture GC cell lines, including 10% FBS and 5% 0.1 mM penicillin-streptomycin in the environment of 5% CO2 at 37 ℃.
Vector construction
Knockdown vectors were generated by using adenovirus following the instructions; GC cells in the exponential growth period were collected and infected with the adenovirus vector containing Eag1 short-hairpin RNAs (shRNAs) or an empty vector. Western blotting was conducted to validate whether the knockdown of Eag1 and empty vector were satisfactory, and then fresh medium was provided for stable cell culture. The sequences for the Eag1 shRNAs were as shown: target 1 (GCGGTCCAATGATACTAAT), target 2 (CCATCTTGGTCCCTTATAA), and target 3 (CAAACTTATCCGCATGAAC).
RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)
Total cell RNA was extracted according to manufacturer’s instructions of TRIzol reagent (Invitrogen, Shanghai, China). Reverse transcription of total RNA was conducted under standard conditions in 10 µl volume with PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). SYBR Green real-time polymerase chain reaction (PCR) was used to measure gene expression of Eag1. The messenger RNA (mRNA) expression level of Eag1 was estimated by the 2×2-ΔΔCt method, taking glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a reference. The specific primer sequences of Eag1 were as follows: (F) 5'-AGAACGTGGATGAGGGCATCAG-3', (R) 5'-ACTGGTAAGGGGTGCCAATG-3'. The primers for GAPDH were (F) 5'-TCAAGAAGGTGGTGAAGCAGG-3' and (R) 5'-TCAAAGGTGGAGGAGTGGGT-3'. Quantitative (q)PCR assays and a LightCycler 480 instrument (Roche, Basel, Switzerland) were utilized to collect the data.
Protein extraction and western blotting
The GC cell lines cultured in the logarithmic growth phase were collected for protein extraction. After transfection for 48 hours, GC cells were treated with cell lysis buffer for 30 minutes and boiled for 10 minutes for denaturation. Electrophoresis in 12% polyacrylamide gels was performed with 40 µg of protein in each sample. In this way, the protein was successfully transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Burlington, MA, USA) and then soaked in tris-buffered saline with Tween 20 (TBST) blocking buffer including 5% skim milk powder and blocked for 2 hours. Antibodies against Eag1, GAPDH, cyclin D1 and cyclin E were incubated overnight at 4 ℃. With washing 5 times in TBST, PVDF membranes were then incubated in the secondary antibody solution with a dilution of 1:50,000 for 2 hours at 37 ℃. An electrochemiluminescence (ECL) detection kit was used to detect the immunoreactive protein bands. The gray values of protein bands were scanned and analyzed by Azure c300 Gel Imaging System (Azure Biosystems, Dublin, USA). Each independent experiment was repeated 3 times.
Cell proliferation and cell colony
Cell lines cultured in logarithmic growth phase were inoculated in 96-well plates. After culturing for 12 hours with serum-free 1640 medium, the GC cells were transfected with Eag1-shRNA and Eag1-shRNA-NC (empty vector). The transfected cells were mixed with 10 µL of cholecystokinin octapeptide (CCK-8; Biosharp, Hefei, China) solution and incubated for another 4 hours at 37 ℃. A microplate reader with the absorbance of 450 nm was used for cell detection.
Clone formation was performed in transfected cells (Eag1-shRNA and empty vector), which were cultured for 3 weeks at 37 ℃ in the presence of 5% CO2. After dropping the culture solution and fixing with 4% methanol for 15 minutes, the plates were stained with iodonitrotetrazolium (INT; Sigma, St. Louis, MO, USA). The number of colonies was counted by microscopic observation. Cloning efficiency was defined as the number of cell colonies/inoculated cell number ×100%.
Cell apoptosis and cell-cycle progression
Cell lines were transfected for 48 hours with Annexin V-APC and 7-AAD and then collected for testing. The information of cell apoptosis and cell cycle progression was measured by flow cytometry [Becton, Dickinson, and Co. (BD) Biosciences, Franklin Lakes, NJ, USA]. Cell Quest software (BD Biosciences) was used to evaluate the time phase distribution of the cell cycle. Cells were divided into 3 types (viable cells, apoptotic cells, and dead cells) according to their state. Each sample was assayed 3 times.
mRNA and protein expression validation
The difference of Eag1 gene expression in tumor sample and normal sample was validated in The Cancer Genome Atlas (TCGA) database. Immunohistochemistry information was provided by The Human Protein Atlas (HPA), in both tumor sample and adjacent normal sample of GC cases. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Functional enrichment analysis
A Volcano plot and heatmap were used to evaluate the differentially expressed genes (DEGs) of Eag1. Functional enrichment analysis including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used for molecular biological functions estimation. Biological processes (BPs), cellular components (CCs), and molecular functions (MFs) were assessed respectively.
Relationship between Eag1 expression and immune infiltration
The relationship between Eag1 expression level and immune infiltration in cancer-associated fibroblasts, T cells, and neutrophils was analyzed by Tumor Immune Estimation Resource version 2 (TIMER2; http://timer.cistrome.org/). The Tumor-Immune System Interaction Database (TISIDB; http://cis.hku.hk/TISIDB/) was conducted to estimate the relationship between Eag1 expression level and abundance of immune cells. Spearman correlation coefficient was calculated for evaluation.
Statistical analysis
The characteristics of continuous variables were presented as the mean ± standard deviation (SD). To compare the difference between two groups, either Student’s t-test or Mann-Whitney U test was performed. One-way analysis of variance (ANOVA) was analyzed to calculate difference of means among more than two groups. Kaplan-Meier survival analysis was used for subgroup analysis of Eag1 gene expression level and GC prognosis. Then, a nomogram was performed with Eag1 expression and some clinicopathological variables. A two-sided P value less than 0.05 was defined as a statistically significant difference. Graphics were drawn by R software (R 3.6.1; The R Foundation for Statistical Computing, Vienna, Austria) and GraphPad Prism 5 software (GraphPad Software, San Diego, CA, USA). All statistical analyses were conducted with SAS 9.4 software (SAS Institute, Cary, NC, USA).
Results
Effects of shRNA treatment on Eag1 expression in GC cells
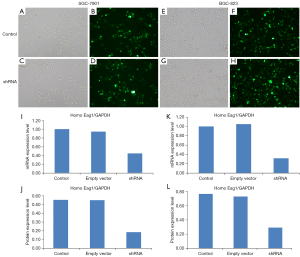
We analyzed the impact of shRNA transfection on mRNA and protein expression of Eag1 in the shRNA vector, empty vector, and control groups. The transfection efficiency of Ad5-Eag1-shRNA in SGC-7901 and BGC-823 are shown in Figure 1A-1H. When compared with control group, the Eag1 gene expression was downregulated by 55.7% in SGC-7901 and 68.3% in BGC-823 in the shRNA group. Similarly, the protein expression of Eag1 was significantly downregulated by 66.7% in the SGC-7901 and downregulated by 61.6% in the BGC-823 transfected with shRNA vector compared to the control group (Figure 1I-1L). The same results were also observed between cell lines transfected with the shRNA vector and empty vector.
Eag1 knockdown and cell proliferation
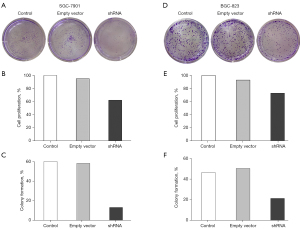
To estimate influence of transfection with Ad5-Eag1-shRNA on cell proliferation, CCK-8 was performed to analyze the rate of cell proliferation and colony formation in different cell groups. The transfection efficiency of Ad5-Eag1-shRNA on SGC-7901 cell proliferation is shown in Figure 2A. As shown in Figure 2, in comparison with the empty vector group and control group, the cell proliferation rate was lower in SGC-7901 transfected with Ad5-Eag1-shRNA (all P<0.05) (Figure 2B). Moreover, the clone formation number in the shRNA group was decreased compared with the empty vector group and control group, and the differences were statistically significant in SGC-7901 cell lines (Figure 2C). Similar pictures were found in BGC-823 cell lines (Figure 2D-2F).
Eag1 knockdown and cell cycle of GC
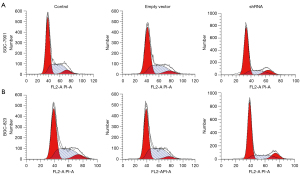
Flow cytometry was performed for the estimation of cell cycle progression as well as the effects of Eag1 knockdown on GC cell cycle. Figure 3 shows that compared with the control group, the cells transfected with Ad5-Eag1-shRNA presented a cell cycle increase at G1-G0 phase (65.7% at 33.73 with shRNA vector vs. 55.5% at 38.25 with the control group for SGC-7901 and 64.8% at 38.89 with shRNA vector vs. 51.7% at 38.68 with the control group for BGC-823). Decreased G2-S phase was also seen in the comparison between GC lines transfected with Ad5-Eag1-shRNA and the control group. Moreover, both SGC-7901 and BGC-823 in the shRNA group and empty vector showed a similar cell cycle effect (Figure 3).
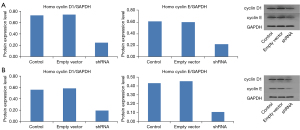
Eag1 knockdown and cyclin D1/cyclin E expression in GC cells
The influence of Eag1 knockdown on protein expression of cyclin D1 and cyclin E was also evaluated. As shown in Figure 4, when GC cells were transfected with Ad5-Eag1-shRNA, cyclin D1 expression was inhibited by 66.3% in the SGC-7901 and 65.4% in the BGC-823 in comparison with control group. Similarly, protein expression of cyclin E was significantly downregulated by 64.8% in the SGC-7901 and downregulated by 74.8% in the BGC-823 transfected with the shRNA vector. The same results were also observed between cell lines transfected with shRNA vector and those with the empty vector (Figure 4).
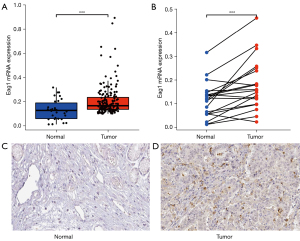
mRNA and protein expression validation
The mRNA expression difference of Eag1 in tumor samples and adjacent normal tissue samples were evaluated in TCGA database. The mRNA expression of Eag1 was higher in tumor samples than in normal samples (0.21±0.12 vs. 0.13±0.08, P<0.001, Figure 5A). The same results were yielded in the difference analysis between tumor samples and adjacent normal samples (0.18±0.11 vs. 0.11±0.07, P<0.001, Figure 5B). Immunohistochemistry information of Eag1 in HPA indicated that the protein level of Eag1 was higher in tumor samples compared with normal samples (Figure 5C,5D).
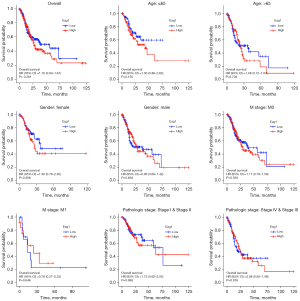
Association between Eag1 expression and GC prognosis
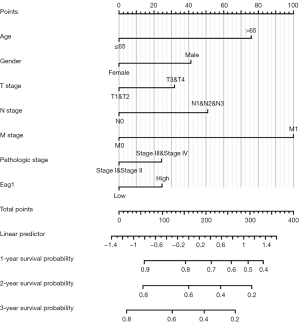
Figure 6 shows the results of Kaplan-Meier survival analysis for subgroup analysis of Eag1 gene expression level and GC prognosis, indicating that patients with higher Eag1 expression may have a less favorable prognosis than overall GC and different characteristics patients (age, gender, M stage, pathologic stage, and so on).
A nomogram was further performed for GC prognosis with Eag1 expression and some common clinicopathological variables according to the results of univariate Cox regression analysis (Figure 7). The 1-, 2-, and 3-year survival probability for GC patients was estimated by combining all the variables in the nomogram. The C-statistic value for nomogram verification was 0.814, suggesting that the nomogram has a satisfactory accuracy for prediction of GC prognosis.
Functional enrichment analysis
Functional enrichment analysis was conducted for the DEGs of Eag1. Genes were divided into two groups with the median of Eag1, and the DEGs of Eag1 were defined as: false discovery rate (FDR) <0.05 and |log2 fold change| >1.5. Finally, 3,388 genes with 3,366 up-regulation genes and 22 down-regulation genes were identified, as shown in Figure 8A. Figure 8B displays the heatmap of the top 10 DEGs.
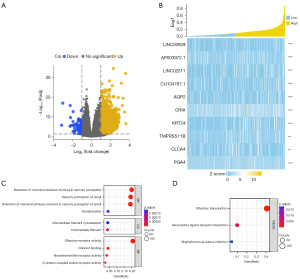
For enrichment analysis, GO and KEGG analyses were conducted for molecular biological functions evaluation (Figure 8C,8D). For the 3,388 DEGs of Eag1, the following BPs were enriched: keratinization, sensory perception of smell, and serotonin receptor signaling pathway. Also, the following CCs were enriched: intermediate filament and intermediate filament cytoskeleton. Lastly, the following MFs were enriched: olfactory receptor activity, G protein-coupled amine receptor activity, and neurotransmitter receptor activity.
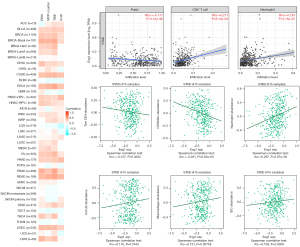
Relationship between Eag1 gene expression and status of immune infiltration
It has been reported that cancer-associated fibroblasts may have a crucial effect on the regulation of tumor infiltrating immune cells (17). In addition, considering the role of Eag1 in actin cytoskeleton structure regulation, the relationship between Eag1 gene expression and status of tumor immune infiltration was further estimated. The EPIC, MCP-counter, TIDE, and Xcell algorithms were employed to evaluate the correlation between Eag1 expression and immune infiltration of cancer-associated fibroblast based on TCGA database, and Eag1 gene expression was strongly related with the status of immune infiltration in bladder cancer (BLCA), breast cancer (BRCA), stomach adenocarcinoma (STAD), and other tumors (Figure 9). We used TISIDB to estimate relationship between Eag1 and status of immune infiltration in GC. It revealed that Eag1 expression was significantly associated with abundance of Tcm-CD4+ T cell, CD56dim, neutrophils, macrophages, and other immune cells (all P<0.05, Figure 9).
Discussion
In this study, according to functional experiment in vitro, we found that the gene and protein expression of Eag1 was downregulated upon transfection with shRNA vector. Knockdown of Eag1 inhibited the GC cell proliferation, whereas knockdown of Eag1 decreased G2-S phase cell cycle progression. Our study estimated the influence of the Eag1 channel on GC progression, indicating that Eag1 knockdown can down-regulate cyclin D1 and cyclin E expression in vitro. This study suggests that the expression of Eag1 may make a critical difference in the occurrence and progress of GC.
Bioinformatic analysis was also analyzed for Eag1 validation with multiple public databases. Significant difference expression of Eag1 gene and protein was observed between tumor samples and normal tissue samples. Kaplan-Meier survival analysis showed that Eag1 expression was strongly related to GC prognosis; the nomogram indicated a satisfied accuracy for prediction of GC prognosis. Enrichment analysis showed that Eag1 and the DEGs can influence the regulation of intermediate filament, intermediate filament cytoskeleton, and G protein-coupled amine receptor activity. In addition, a significant correlation between Eag1 expression and GC immune infiltration was seen based on TISIDB and TIMER.
As a kind of voltage-gated potassium channel, the Eag gene was first reported in 1969 and was shown to cause Drosophila melanogaster to move slowly and regularly under anesthesia (18). Eag1 has been proved to be an important member of Eag subfamily, and can encode a member of subfamily H, which can affect the normal physiological activity of potassium channels by regulating voltage gating (19). Pardo et al. confirmed the close relationship between Eag1 and the development of tumors for the first time in 1999 (20). Several studies have shown that an abnormal expression state of Eag1 in human tumor cell lines and tumor tissue can influence the proliferation, invasion, and other biological functions of tumor cells (21-23). This suggests that Eag1 may be a useful tumor biomarker for therapy in different cancers. The Eag1 channel consists of 6 transmembrane helices, including potassium selective pores and voltage sensors (24). Studies have shown that the voltage sensor in the Eag1 channel may undergo structural changes and then regulate the channel switch when the membrane potential changes (25-27). As an Eag1 channel inhibitor, astemizole may be a potential drug to restrain the proliferation of tumor cells because of its target proteins associated with cancer (28). Some studies have found that cell proliferation is blocked when measured with astemizole, suggesting that astemizole may play a crucial role in Eag1 channel inhibition (29,30). A similar conclusion was obtained in an in vivo study (31). However, the role of the Eag1 channel in possible biological mechanism of GC remains unclear.
This study showed Eag1 expression may have a certain influence on the progression of GC through regulating cyclin D1 and cyclin E expression according to functional experiments. However, there are still limitations to the study. Only the effect of Eag1 in GC development in tumor cell lines was considered. However, some studies have confirmed a strong relationship between Eag1 expression and GC development in animal models (32,33). In consideration of the functional experimental results presented in this study, our findings still provide important information regarding the Eag1 channel and the initiation and development of GC. Further studies, including experiments in animal models and human populations, are planned to validate the impact of Eag1 on the development of and susceptibility to GC.
Conclusions
In conclusion, this study found that the expression of Eag1 may affect the initiation and occurrence of GC through regulating cyclin D1 and cyclin E expression. Further functional experiments including animal models and population studies with sufficient samples are necessary to verify our findings.
Acknowledgments
We are grateful to all the public databases (TCGA, HPA, TIMER2 and TISIDB) for providing all the data used in this study.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2276/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2276/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2276/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol 2020;18:534-42. [Crossref] [PubMed]
- Zhao Y, Zhang J, Cheng ASL, et al. Gastric cancer: genome damaged by bugs. Oncogene 2020;39:3427-42. [Crossref] [PubMed]
- Docampo E, Martin M, Gangolf M, et al. Heredity and cancer. Rev Med Liege 2021;76:327-36. [PubMed]
- Petryszyn P, Chapelle N, Matysiak-Budnik T. Gastric Cancer: Where Are We Heading? Dig Dis 2020;38:280-5. [Crossref] [PubMed]
- Pardo LA, Stühmer W. Eag1: an emerging oncological target. Cancer Res 2008;68:1611-3. [Crossref] [PubMed]
- Fang YC, Fu SJ, Hsu PH, et al. Identification of MKRN1 as a second E3 ligase for Eag1 potassium channels reveals regulation via differential degradation. J Biol Chem 2021;296:100484. [Crossref] [PubMed]
- Lin H, Li Z, Chen C, et al. Transcriptional and post-transcriptional mechanisms for oncogenic overexpression of ether a go-go K+ channel. PLoS One 2011;6:e20362. [Crossref] [PubMed]
- Rodríguez-Rasgado JA, Acuña-Macías I, Camacho J. Eag1 channels as potential cancer biomarkers. Sensors (Basel) 2012;12:5986-95. [Crossref] [PubMed]
- Ding XW, Luo HS, Jin X, et al. Aberrant expression of Eag1 potassium channels in gastric cancer patients and cell lines. Med Oncol 2007;24:345-50. [Crossref] [PubMed]
- Li Z, Zhu K, Gong X, et al. Inducing Polyclonal Eag1-Specific Antibodies by Vaccination with a Linear Epitope Immunogen and Its Relation to Breast Tumorigenesis. Pathol Oncol Res 2017;23:761-7. [Crossref] [PubMed]
- Pardo LA, Contreras-Jurado C, Zientkowska M, et al. Role of voltage-gated potassium channels in cancer. J Membr Biol 2005;205:115-24. [Crossref] [PubMed]
- Chen J, Xuan Z, Song W, et al. EAG1 enhances hepatocellular carcinoma proliferation by modulating SKP2 and metastasis through pseudopod formation. Oncogene 2021;40:163-76. [Crossref] [PubMed]
- Chávez-López MG, Zúñiga-García V, Castro-Magdonel BE, et al. Eag1 Gene and Protein Expression in Human Retinoblastoma Tumors and its Regulation by pRb in HeLa Cells. Genes (Basel) 2020;11:119. [Crossref] [PubMed]
- Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov 2019;18:99-115. [Crossref] [PubMed]
- Kaplan WD, Trout WE 3rd. The behavior of four neurological mutants of Drosophila. Genetics 1969;61:399-409. [Crossref] [PubMed]
- Wang X, Chen Y, Li J, et al. Tetrandrine, a novel inhibitor of ether-à-go-go-1 (Eag1), targeted to cervical cancer development. J Cell Physiol 2019;234:7161-73. [Crossref] [PubMed]
- Pardo LA, del Camino D, Sánchez A, et al. Oncogenic potential of EAG K(+) channels. EMBO J 1999;18:5540-7. [Crossref] [PubMed]
- Toplak Ž, Hendrickx LA, Abdelaziz R, et al. Overcoming challenges of HERG potassium channel liability through rational design: Eag1 inhibitors for cancer treatment. Med Res Rev 2022;42:183-226. [Crossref] [PubMed]
- Asher V, Khan R, Warren A, et al. The Eag potassium channel as a new prognostic marker in ovarian cancer. Diagn Pathol 2010;5:78. [Crossref] [PubMed]
- Shi Y, Yang X, Xue X, et al. HANR promotes lymphangiogenesis of hepatocellular carcinoma via secreting miR-296 exosome and regulating EAG1/VEGFA signaling in HDLEC cells. J Cell Biochem 2019;120:17699-708. [Crossref] [PubMed]
- Whicher JR, MacKinnon R. Regulation of Eag1 gating by its intracellular domains. Elife 2019;8:49188. [Crossref] [PubMed]
- Wynia-Smith SL, Gillian-Daniel AL, Satyshur KA, et al. hERG gating microdomains defined by S6 mutagenesis and molecular modeling. J Gen Physiol 2008;132:507-20. [Crossref] [PubMed]
- Thouta S, Sokolov S, Abe Y, et al. Proline scan of the HERG channel S6 helix reveals the location of the intracellular pore gate. Biophys J 2014;106:1057-69. [Crossref] [PubMed]
- Wang X, Chen Y, Liu H, et al. A novel anti-cancer mechanism of Nutlin-3 through downregulation of Eag1 channel and PI3K/AKT pathway. Biochem Biophys Res Commun 2019;517:445-51. [Crossref] [PubMed]
- García-Quiroz J, Camacho J. Astemizole: an old anti-histamine as a new promising anti-cancer drug. Anticancer Agents Med Chem 2011;11:307-14. [Crossref] [PubMed]
- Gómez-Varela D, Zwick-Wallasch E, Knötgen H, et al. Monoclonal antibody blockade of the human Eag1 potassium channel function exerts antitumor activity. Cancer Res 2007;67:7343-9. [Crossref] [PubMed]
- de Guadalupe Chávez-López M, Pérez-Carreón JI, Zuñiga-García V, et al. Astemizole-based anticancer therapy for hepatocellular carcinoma (HCC), and Eag1 channels as potential early-stage markers of HCC. Tumour Biol 2015;36:6149-58. [Crossref] [PubMed]
- Ufartes R, Schneider T, Mortensen LS, et al. Behavioural and functional characterization of Kv10.1 (Eag1) knockout mice. Hum Mol Genet 2013;22:2247-62. [Crossref] [PubMed]
- Asher V, Sowter H, Shaw R, et al. Eag and HERG potassium channels as novel therapeutic targets in cancer. World J Surg Oncol 2010;8:113. [Crossref] [PubMed]
- Weber C, de Queiroz FM, Downie BR, et al. Silencing the activity and proliferative properties of the human EagI Potassium Channel by RNA Interference. J Biol Chem 2006;281:13030-7. [Crossref] [PubMed]