
Comprehensive bioinformatics analysis for MEF2 family genes in gastric cancer
Introduction
Gastric cancer (GC) is the third most common cause of cancer-related death, and about 784,000 cancer-related deceases are caused because of GC in 2018 all around the world (1). Despite the progress in early discovery, radical therapy operation, and multimodal cure modalities, it remains difficult to cure GC, and patients had a poor prognosis with a median overall survival of 1 year for advanced status in Western countries(2,3). the postoperative relapse or metastasis often happens after radical operation in 40–60% of GC patients (4). It is still urgent to improve the clinical outcome of patients with GC and investigate the underlying molecular mechanism of carcinogenesis, which may provide a new strategy to find out the patients with high relapse rates and improve the prognosis.
MEF2 family members act as transcriptional factors, which play a critical role in cellular proliferation, differentiation, apoptosis, and survival (5-7). Increasing evidence suggests that four MEF2 family members (A-D) participated in the pathogenesis and development of cancer by modulating complex protein networks (8,9). Overexpression of MEF2 impacted histone hyperacetylation and immune checkpoint molecules, resulting in the progression of hepatocellular carcinoma (10,11). MEF2C was found to mediate vascular endothelial growth factor (VEGF) induced malignancy enhancement (12). In addition, MEF2D was activated by HIF-1α and involved in colorectal cancer angiogenesis (13). MEF2D was shown to significantly activate the Wnt/β-catenin pathway and contributed to the invasion of GC (8). However, evidence about the role of MEF2 in the pathogenesis of GC is still lacking.
Weighted gene co-expression network analysis (WGCNA) is a systematic biology algorithm, which has been used to examine the connection between gene sets and clinical traits by constructing a gene co-expression network (14). The purpose of this study was to investigate the prognostic significance of MEF2 family members and identify network-centric genes in GC by applying the WGCNA algorithm.
Methods
Data acquisition and preprocessing
Flowchart showed our work content and order (Figure S1). The Cancer Genome Atlas (TCGA) (15) is a well-known public database containing genomic and clinical information of various tumors. We acquired the gene expression matrix (TCGA-STAD.htseq_fpkm.tsv.gz), microRNA (miRNA) expression matrix (TCGA-STAD.mirna.tsv.gz), clinical information (TCGA-STAD.GDC_phenotype.tsv.gz), copy number variation file (TCGA-STAD.gistic.tsv.gz), and DNA methylation matrix (TCGA-STAD.methylation450.tsv.gz) of GC patients through the UCSC Xena website (https://xena.ucsc.edu/) (16). Single nucleotide polymorphism (SNP) file “gdc_download_20211224_143028.203685.tar.gz” were obtained through https://portal.gdc.cancer.gov/. GSE84434 containing transcriptome and clinical data of GC were downloaded from the Gene Expression Omnibus (GEO) website (https://www.ncbi.nlm.nih.gov/gds). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The gene expression values of the transcriptome matrix were transformed into a log2 form for further analyses. The expression levels of MEF2A, MEF2B, MEF2C, and MEF2D were extracted, and we conducted principal component analysis (PCA) for four genes expression to obtain PCA1 regarded as the levels of the MEF2 family.
Considering the nature of DNA methylation array, we defined beta value <0.3 as unmethylated, 0.3 ≤ beta value <0.6 as partial methylation, and beta value ≥0.6 as complete methylation. Integer 0, 1, 2 were used to measure the three status unmethylated, partial methylation, and complete methylation.
Differential expression and prognostic analysis
UALCAN (http://ualcan.path.uab.edu/analysis.html) and GEPIA2 (http://gepia2.cancer-pku.cn/#index) are online bioinformatics analyses tools for TCGA data (17,18). The differential analysis aims to screen out genes or miRNAs with distinct distribution between tumor and normal samples; the absolute value of logFC >0.5 and FDR <0.05 means significant. The differential expression analysis of MEF2 family genes and 7 miRNA regulating genes were conducted using GEPIA2, and those of miRNAs were performed in UALCAN. Besides, Kaplan-Meier overall survival analyses for miRNAs and genes regulating those miRNAs were conducted using the survival package in the R language. The surv cutpoint function was applied to calculate the optimal cut-off that divided the samples into the high-expression and low-expression groups on the genes. P value <0.05 was considered significant.
Establishment of WGCNA and identification of modules
WGCNA was performed to acquire MEF2 family related modules and hub genes using the WGCNA package in R language. Before conducting WGCNA, we divided the GC samples according to the quartile of MEF2 expression levels; samples with MEF2 expression levels in the upper quartile were considered as the high-expression group (94 samples), and with MEF2 expression levels in the last quartile were regarded as the low-expression group (94 samples).
The establishment of co-expression network needs the following primary steps: (I) Calculate the similarity matrix; (II) Choose the weighting coefficient β, and convert the similarity matrix into an adjacency matrix; (III) Turn the adjacency matrix into a TOM (topology overlap matrix); (IV) Conduct the hierarchical clustering for dissimilarity (1-TOM) to acquire the hierarchical clustering tree; (V) Identify the modules from the hierarchical clustering tree using the dynamic tree cut method and (VI) Compute the module eigengene (ME) of modules. The ME means the overall expression level of the module. The Pearson correlation coefficients between the MEs of the modules were computed, and the average distance between the MEs of all modules was calculated using the 1-Pearson correlation coefficient. The average-linkage hierarchical clustering method based on a minimum size (gene group) of 50 was applied to group the MEs of all modules. The modules with high similarity were combined to obtain the co-expression network.
However, genes with a weak correlation with module membership (MM) in the modules would affect the results of the module cluster. Iterative WGCNA was used to figure out the problem. Genes with MM >0.6 were extracted and analyzed by WGCNA again; as for mRNA analyses, we performed twice.
We picked MEF2 related modules according to the following criteria: (I) the ME of modules owns a high correlation with MEF2 levels; (II) the module’s average absolute value of gene significance (GS) is relatively higher; (III) GS had a high correlation with MM.
Genes highly interconnecting with the modules’ nodes were considered the hub genes, representing functional importance. The networkScreening function of the WGCNA package according to GS and MM was conducted to screen out hub genes directly. Genes with q-weighted cutoff <0.001 were selected as hub genes.
Enrichment analyses
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) were chosen to evaluate hub genes’ biological functions and signalling pathways. The clusterProfiler package was used to implement these analyses. The terms of GO with q value <0.001 and that of KEGG with q value <0.01 were considered significant, respectively (19,20).
Identification of MEF2 related miRNAs
KEGG enrichment analyses showed that hub genes in the green module were involved in “miRNAs in cancer”. We extracted these 9 hub genes in the “miRNAs in cancer” pathway. Subsequently, the expression matrix of 9 hub genes and miRNAs were merged for WGCNA, through which we could identify the MEF2 related miRNAs which were regulated by the 9 hub genes. TARGETSCAN websites (http://www.targetscan.org/vert_80/) were employed to validate the miRNAs targeting MEF2 family members (21). Cytoscape (version 3.8.2) was used to make up the interaction network (22).
Statistical analysis
All analyses in this study were conducted using the R language (version 4.0.5). P<0.05 was considered statistically significant.
Results
MEF2 family members were dysregulated in GC patients
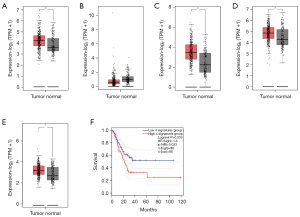
We respectively examined the prognostic values of the mRNA expression of 4 MEF2 family members in GC patients in GEPIA (www.cancer-pku.cn). Compared with normal stomach tissue, MEF2A, MEF2C, and the MEF2D expression level were significantly higher in GC tissue, while the expression level of MEF2B showed no statistically significant difference between normal tissue and GC tissue (Figure 1). We further compared the expression level of MEF2 family members on set-level. The MEF2 family members expressed higher in GC tissue as a gene set than in normal tissue (Figure 1E). In addition, we divided 192 GC patients into two groups according to the expression levels of MEF2 family members. We found out that GC patients of the high 4 signature group had a significantly worse survival than patients of the low 4 signature group (P=0.033, Figure 1F), which revealed the negative influence of the MEF2 family on the outcome of GC patients.
MEF2 genes represent “preferences” in some histological types
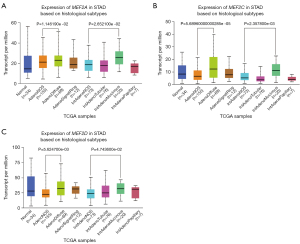
GC exhibits various histological types like adenocarcinoma type and intestinal adenocarcinoma type. We wonder whether regular patterns exist between MEF2 genes and some histological types. According to the expression of MEF2 genes in STAD based on histological types, we found that three MEF2 genes except for MEF2B exhibit higher expression in Adenocarcinoma diffuse type and Intestinal adenocarcinoma Mucinous type compared with the corresponding not otherwise specified (NOS) type (Figure 2).
Genomic variation of MEF2 genes in GC
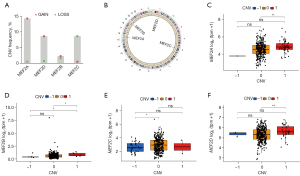
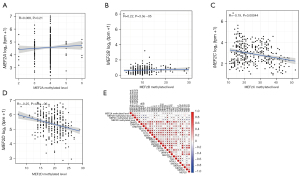
Copy number variation (CNV) and SNP are frequent variable types for most cancer-related genes, so do MEF2 genes. CNV frequency analysis showed that 14.32%, 2.27%, 0.46%, and 8.64% of GC patients exhibited copy number gain of MEF2A, MEF2B, MEF2C, and MEF2D, respectively. as for copy number loss, MEF2A, MEF2B, MEF2C, and MEF2D correspond to 0.46%, 1.6%, 8.64%, and 0.68% (Figure 3A). The overall copy number level for MEF2 genes is copy number gain except for MEF2C with copy number loss (Figure 3B). In most cases, increasing copy number may enhance the transcriptome level, and MEF2 also follow this law. The differential expression analysis showed that MEF2 family genes expression increased significantly as the copy number rose (Figure 3C-3F). Even if the remarkable variance of copy number for MEF2 genes, they exhibit a low frequency of SNP (Figure S2). Furthermore, correlation analysis between the DNA methylation and transcriptome expression of four MEF2 genes were conducted (Figure 4A-4D), the results of which showed high DNA methylation along with low expression of MEF2C and MEF2D, and MEF2A and MEF2B expression positively correlated with DNA methylation level. We also evaluate the correlation between DNA methylase genes expression and the overall methylation level of four MEF2 genes, and found that the overall DNA methylation level of four MEF2 genes simultaneously correlated with DNMT1 and UHRF1 (Figure 4E).
Construction of co-expression module of GC and identification of critical modules
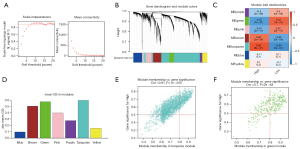
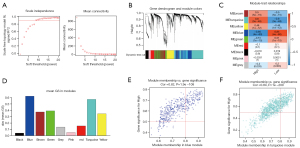
In our study, we applied iterative WGCNA to decrease the noise genes. In the TCGA STAD cohort, the power of β=7 (scale-free R2=0.9) as the soft thresholding ensures a scale-free topology model (Figure 5A,5B). A total of 7 modules were identified (Figure 5C). The relevance between modules and MEF2 expression levels was tested using three methods. Firstly, the turquoise and green modules had the highest MEs (Figure 5C). Secondly, the turquoise and green modules had the highest average GS (Figure 5D). Lastly, GS correlated with MM in the turquoise module and the green module (Figure 5E,5F). Thus, we identified that the turquoise and green modules were the most relevant to the MEF2 family. Afterward, we also identified two vital MEF2-related blue and turquoise modules, using the same method in another GC cohort, GSE84434 (Figure 6).
Identification of hub genes and functional enrichment analysis
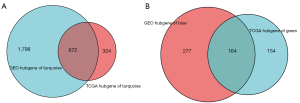
Furthermore, hub genes within each module were identified. Hub genes screening was required to meet three conditions. The first was that the hub genes were in the modules (green and turquoise modules); the second was that the hub genes met the screening criteria (q-weighted <0.001) of the ‘networkScreening’ function of the WGCNA method. As for TCGA modules, 996 and 318 hub genes were extracted in the turquoise and green module, respectively. GSE84434 modules turquoise and blue included 2470 and 441 hub genes, respectively. Moreover, we found that TCGA turquoise and GSE84434 turquoise module are homogeneous, and so are TCGA green and GSE84434 blue modules. The two homogeneous modules contain a large proportion of shared genes (Figure 7). Two sets of overlap hub genes were obtained after the intersection. Those from TCGA green and GSE84434 blue modules were defined as “set A”, while “set B” overlap hub genes were obtained from turquoise modules.
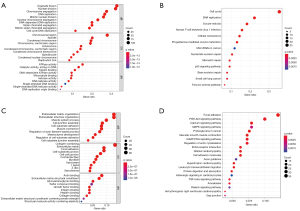
To investigate the potential mechanisms of MEF2 family in GC, functional enrichment analyses and pathway enrichment analyses were applied. As for “set A”, Enrichment analysis showed that genes of set A mainly related to intranuclear activity and cell cycle (Figure 8A,8B). While set B genes primarily enrich in extranuclear activity and signal transduction (Figure 8C,8D). As a result, MEF2 related gene set can be divided into two sections: set A is an intranuclear set, and set B belongs to an extranuclear group.
Identification of MEF2 related miRNAs
One of the enriched pathways of hub genes in set A was miRNAs in cancer. These hub genes were identified as miRNA modulators, including cell division cycle 25 (CDC25) A-C, cell division cycle associated 5 (CDCA5), E2F1, E2F2, cyclin E1 (CCNE1), CCNE2, enhancer of zeste homologue 2 (EZH2), stathmin 1 (STMN1), breast cancer 1 (BRCA1), kinesin super family 23 (KIF23) and DNA methyltransferase 1 (DNMT1). These modulators were combined with the miRNAs expression matrix, and co-expression modules were constructed using WGCNA. The most significant module was the turquoise module (Figure S3). Furthermore, hub genes within the turquoise module were identified, including 25 miRNAs and 7 modulators.
The expression levels related to these miRNAs and modulator genes were investigated. All 25 miRNAs and 7 miRNA modulators were significantly upregulated in GC tissues compared with normal tissues, as summarized in Table S1 and Figure S4. We further investigated overall survival related to these genes. High expression levels of CDC25A, CDCA5, E2F1, EZH2, and KIF23 correlated with better survival of GC patients (Figure S5A-S5E). miRNA-7-1, miRNA-17, miRNA-183, and miRNA-942 had a positive impact on the survival of GC patients; Whereas miRNA-210 and miRNA-219a-1 had a negative impact on the survival (Figure S5F-S5K). Since high expression levels of these miRNAs were associated with low expression levels of MEF2 family members (Figure S6A), it was presumed that they could target members of the MEF2 family. The transcriptional regulatory network was demonstrated with verification on the Targetscan website, as shown in Figure S6B.
Discussion
In our study, we profile the four MEF2 genes in GC by integrating analyzing epigenetic, genetic, and transcriptome data. Consistent with previous studies of different tumor types (10,23,24), we found that MEF2 family members were overexpressed in GC tissues. However, the underlying mechanisms have not been fully illustrated.
MEF2A, MEF2C, and MEF2D exhibit higher expression in some histological types of GC, including adenocarcinoma diffuse type and intestinal Adenocarcinoma Mucinous type compared with their NOS type. Some researchers have reported MEF2 function in histological formation Vincentz et al. found that Nkx2.5 interacting with MEF2C may be essential for ventricle formation (25). Besides, Salivary gland adenocarcinoma with MEF2C-SS18 fusion exhibited distinct histological performance (26). These pieces of evidence support the hypothesis that MEF2 genes are involved in histological change in GC.
CNV gain of proliferation-related genes results from evolutionary selection during cancer development (27). CNV also affect the protein-coding and non-coding RNA level (28). We found that MEF2 family genes exhibit remarkable copy number change, especially MEF2A, MEF2C, and MEF2D. Meanwhile, MEF2 genes transcriptome level rose as the copy number increased. Copy number variance represents an essential factor for MEF2 gene expression. CNV of MEF2 genes may be a potential biological target for GC curation.
Given epigenetic alterations have emerged as potential clinical biomarkers indicating gastrointestinal cancer diagnosis and therapy. DNA methylation of CpG islands may affect the gene expression by regulating the promotor, a critical mechanism that enhances oncogene expression (29). We calculated the overall DNA methylation level of four MEF2 genes, three genes MEF2B, MEF2C, and MEF2D, exhibited a relatively high level of methylation status compared with MEF2A. In addition, correlation analysis showed high DNA methylation along with low expression of MEF2C and MEF2D, while MEF2A and MEF2B expression positively correlated with DNA methylation level. The contrast is worth forward investigating. DNMT1 is a DNA methylase functioning in maintaining DNA methylation during the replication of DNA, and we find that the overall DNA methylation level of four MEF2 genes simultaneously correlated with DNMT1 and a methylation reader UHRF1. DNMT1 and UHRF1 may play a vital role in the DNA methylation of MEF2 genes.
WGCNA was applied to screen out two MEF2 related modules by combining two GC cohorts marked as “set A” and “set B”. According to the GO and KEGG enrichment analysis, genes in set A are mainly involved in intranuclear regulation such as cell cycle, DNA replication, MicroRNA regulation, and biology process within chromosomes. The MEF2 family was DNA-binding transcription factors, and set A represented the “intranuclear set”. In contrast, the set B, an “extracellular set”, is primarily associated with extracellular signal transduction regulation, including the calcium signalling pathway, PI3K-AKT signalling pathway, etc. MEF2 regulates cell cycle, differential and death via receiving signals from the Calcium dependence signalling pathway, including MAPK, PI3K-AKT and TGF-beta signalling pathway (30) Little literature has reported the role of the above pathway in GC through the MEF2 family. We discovered that those signaling transduction pathways associated with the MEF2 family affect GC patients’ prognosis. The further experiment is needed to dig into the deep underlying mechanism.
Six miRNAs (miRNA-7-1, miRNA-17, miRNA-183, miRNA-210, miRNA-219a-1, and miRNA-942) and 5 miRNA modulators (CDC25A, CDCA5, E2F1, EZH2, and KIF23) were found to be significantly correlated with MEF2 expression and have a prognostic impact on GC patients. CDC25A is a member of the cell division cycle 25 family, which has been reported to be overexpressed in a wide range of cancer types and associated with cancer cell survival and tumor growth (31-34). Unlike some previous studies (35-37), we found that overexpression of CDC25A could prolong GC survival. CDCA5 was first identified as a sister chromatin cohesion regulator (38). Only a few investigations implied CDCA5 enhanced proliferation and migration of GC cell lines (39,40). Our study revealed that CDCA5 acted as a protective factor for the overall survival of GC patients. As a member of the E2F family, E2F1 plays a critical role in cell cycle regulation (41). Numerous investigations have reported the mechanisms of how E2F1 promotes tumorigenesis, such as maintaining stemness properties (42,43). However, some investigations had conflicting results, indicating E2F1 suppressed gastric tumor cell proliferation (44). The underlying mechanisms were complex and far from fully understood. Our results showed that patients with overexpression of E2F1 had a better prognosis. As a critical epigenetic enzyme, EZH2 was correlated with gastric tumorigenesis (45,46). KIF23 expression was upregulated in GC tissues and promoted the proliferation of GC cells (47). However, according to our survival analyses, these two genes were associated with better survival, which inferred a complex interaction of a multiple-gene network. The underlying mechanisms were worth being further investigated.
miRNA-17 accelerated cancer cell migration and invasion in multiple solid tumors, such as cervical cancer (48) and colorectal cancer (49). However, we found that miRNA-17 might have an opposite role in GC. miRNA-183 was considered to act as an oncogene in many cancer types in vitro (50,51), but the survival analysis indicated that patients with miRNA-183 overexpression had a better prognosis. The prognostic role of miRNA-210 in GC was still not clear. Our results showed miR-210 had a negative impact on GC survival, which might be due to its involvement in multiple biological processes, such as mitochondrial metabolism, angiogenesis, DNA damage, etc. (52). Except for the miRNAs mentioned above, the prognostic significance of miRNA-7-1, miRNA-942 and miRNA-219a-1 was revealed by our study for the first time.
Although this study reveals essential discoveries, there are also limitations. Firstly, it should be noted that this study has been taken place only by using genomic and clinical information from the public database. Secondly, this study identified genes and miRNAs affecting the survival of GC patients, but the co-expression and interactions of these genes with MEF2 family members still need in-depth exploration.
In summary, the present study profile the variance of four MEF2 genes in GC from genomic, epigenomic, and transcriptome aspect. Iterative WGCNA was conducted to identify two sets of MEF2 related hub genes that enrichment analysis separate them into “intranuclear set” and “extracellular set”. By analyzing the “intranuclear set”, we screened out 6 miRNAs and 5 miRNA modulators that co-expressed with the MEF2 family and prognostic significance. Considering the bioinformatics analysis, further investigates are needed for validation and elucidating the deep underlying mechanism.
Acknowledgments
Funding: This research was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-373/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Yang W, Raufi A, Klempner SJ. Targeted therapy for gastric cancer: molecular pathways and ongoing investigations. Biochim Biophys Acta 2014;1846:232-7. [PubMed]
- GASTRIC (Global Advanced/Adjuvant Stomach Tumor Research International Collaboration) Group. Role of chemotherapy for advanced/recurrent gastric cancer: an individual-patient-data meta-analysis. Eur J Cancer 2013;49:1565-77. [Crossref] [PubMed]
- Cao Y, DePinho RA, Ernst M, et al. Cancer research: past, present and future. Nat Rev Cancer 2011;11:749-54. [Crossref] [PubMed]
- Badodi S, Baruffaldi F, Ganassi M, et al. Phosphorylation-dependent degradation of MEF2C contributes to regulate G2/M transition. Cell Cycle 2015;14:1517-28. [Crossref] [PubMed]
- Homminga I, Pieters R, Langerak AW, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 2011;19:484-97. [Crossref] [PubMed]
- Wu J, Kubota J, Hirayama J, et al. p38 Mitogen-activated protein kinase controls a switch between cardiomyocyte and neuronal commitment of murine embryonic stem cells by activating myocyte enhancer factor 2C-dependent bone morphogenetic protein 2 transcription. Stem Cells Dev 2010;19:1723-34. [Crossref] [PubMed]
- Xu K, Zhao YC. MEF2D/Wnt/β-catenin pathway regulates the proliferation of gastric cancer cells and is regulated by microRNA-19. Tumour Biol 2016;37:9059-69. [Crossref] [PubMed]
- Zhu HX, Shi L, Zhang Y, et al. Myocyte enhancer factor 2D provides a cross-talk between chronic inflammation and lung cancer. J Transl Med 2017;15:65. [Crossref] [PubMed]
- Bai X, Wu L, Liang T, et al. Overexpression of myocyte enhancer factor 2 and histone hyperacetylation in hepatocellular carcinoma. J Cancer Res Clin Oncol 2008;134:83-91. [Crossref] [PubMed]
- Xiang J, Zhang N, Sun H, et al. Disruption of SIRT7 Increases the Efficacy of Checkpoint Inhibitor via MEF2D Regulation of Programmed Cell Death 1 Ligand 1 in Hepatocellular Carcinoma Cells. Gastroenterology 2020;158:664-678.e24. [Crossref] [PubMed]
- Bai XL, Zhang Q, Ye LY, et al. Myocyte enhancer factor 2C regulation of hepatocellular carcinoma via vascular endothelial growth factor and Wnt/β-catenin signaling. Oncogene 2015;34:4089-97. [Crossref] [PubMed]
- Xiang J, Sun H, Su L, et al. Myocyte enhancer factor 2D promotes colorectal cancer angiogenesis downstream of hypoxia-inducible factor 1α. Cancer Lett 2017;400:117-26. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [Crossref] [PubMed]
- Haeussler M, Zweig AS, Tyner C, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res 2019;47:D853-8. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- McGeary SE, Lin KS, Shi CY, et al. The biochemical basis of microRNA targeting efficacy. Science 2019;366:eaav1741.
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Song Z, Feng C, Lu Y, et al. Overexpression and biological function of MEF2D in human pancreatic cancer. Am J Transl Res 2017;9:4836-47. [PubMed]
- Zhang S, Li Z, Zhang L, et al. MEF2-activated long non-coding RNA PCGEM1 promotes cell proliferation in hormone-refractory prostate cancer through downregulation of miR-148a. Mol Med Rep 2018;18:202-8. [Crossref] [PubMed]
- Vincentz JW, Barnes RM, Firulli BA, et al. Cooperative interaction of Nkx2.5 and Mef2c transcription factors during heart development. Dev Dyn 2008;237:3809-19. [Crossref] [PubMed]
- Bishop JA, Weinreb I, Swanson D, et al. Microsecretory Adenocarcinoma: A Novel Salivary Gland Tumor Characterized by a Recurrent MEF2C-SS18 Fusion. Am J Surg Pathol 2019;43:1023-32. [Crossref] [PubMed]
- Mauro JA, Butler SN, Ramsamooj M, et al. Copy number loss or silencing of apoptosis-effector genes in cancer. Gene 2015;554:50-7. [Crossref] [PubMed]
- Liang L, Fang JY, Xu J. Gastric cancer and gene copy number variation: emerging cancer drivers for targeted therapy. Oncogene 2016;35:1475-82. [Crossref] [PubMed]
- Grady WM, Yu M, Markowitz SD. Epigenetic Alterations in the Gastrointestinal Tract: Current and Emerging Use for Biomarkers of Cancer. Gastroenterology 2021;160:690-709. [Crossref] [PubMed]
- McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci 2002;27:40-7. [Crossref] [PubMed]
- Wu W, Fan YH, Kemp BL, et al. Overexpression of cdc25A and cdc25B is frequent in primary non-small cell lung cancer but is not associated with overexpression of c-myc. Cancer Res 1998;58:4082-5. [PubMed]
- Al-Matouq J, Holmes T, Hammiller B, et al. Accumulation of cytoplasmic CDC25A in cutaneous squamous cell carcinoma leads to a dependency on CDC25A for cancer cell survival and tumor growth. Cancer Lett 2017;410:41-9. [Crossref] [PubMed]
- Sun Y, Li S, Yang L, et al. CDC25A Facilitates Chemo-resistance in Ovarian Cancer Multicellular Spheroids by Promoting E-cadherin Expression and Arresting Cell Cycles. J Cancer 2019;10:2874-84. [Crossref] [PubMed]
- Chen S, Tang Y, Yang C, et al. Silencing CDC25A inhibits the proliferation of liver cancer cells by downregulating IL-6 in vitro and in vivo. Int J Mol Med 2020;45:743-52. [Crossref] [PubMed]
- Guo SL, Ye H, Teng Y, et al. Akt-p53-miR-365-cyclin D1/cdc25A axis contributes to gastric tumorigenesis induced by PTEN deficiency. Nat Commun 2013;4:2544. [Crossref] [PubMed]
- Wei W, Mo X, Yan L, et al. Circular RNA Profiling Reveals That circRNA_104433 Regulates Cell Growth by Targeting miR-497-5p in Gastric Cancer. Cancer Manag Res 2020;12:15-30. [Crossref] [PubMed]
- Zhao D, Chen H, Wang B. Assessing the Regulatory Functions of LncRNA SNHG11 in Gastric Cancer Cell Proliferation and Migration. Front Cell Dev Biol 2021;9:620476. [Crossref] [PubMed]
- Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell 2005;18:185-200. [Crossref] [PubMed]
- Chen T, Huang Z, Tian Y, et al. Role of triosephosphate isomerase and downstream functional genes on gastric cancer. Oncol Rep 2017;38:1822-32. [Crossref] [PubMed]
- Huang Z, Zhang S, Du J, et al. Cyclin-Dependent Kinase 1 (CDK1) is Co-Expressed with CDCA5: Their Functions in Gastric Cancer Cell Line MGC-803. Med Sci Monit 2020;26:e923664. [Crossref] [PubMed]
- Alonso MM, Fueyo J, Yung WK, et al. E2F1 and telomerase: alliance in the dark side. Cell Cycle 2006;5:930-5. [Crossref] [PubMed]
- Fu Y, Hu C, Du P, et al. E2F1 Maintains Gastric Cancer Stemness Properties by Regulating Stemness-Associated Genes. J Oncol 2021;2021:6611327. [Crossref] [PubMed]
- Enjoji S, Yabe R, Tsuji S, et al. Stemness Is Enhanced in Gastric Cancer by a SET/PP2A/E2F1 Axis. Mol Cancer Res 2018;16:554-63. [Crossref] [PubMed]
- Xiao Q, Li L, Xie Y, et al. Transcription factor E2F-1 is upregulated in human gastric cancer tissues and its overexpression suppresses gastric tumor cell proliferation. Cell Oncol 2007;29:335-49. [PubMed]
- Pan YM, Wang CG, Zhu M, et al. STAT3 signaling drives EZH2 transcriptional activation and mediates poor prognosis in gastric cancer. Mol Cancer 2016;15:79. [Crossref] [PubMed]
- Huang B, Mu P, Yu Y, et al. Inhibition of EZH2 and activation of ERRγ synergistically suppresses gastric cancer by inhibiting FOXM1 signaling pathway. Gastric Cancer 2021;24:72-84. [Crossref] [PubMed]
- Li XL, Ji YM, Song R, et al. KIF23 Promotes Gastric Cancer by Stimulating Cell Proliferation. Dis Markers 2019;2019:9751923. [Crossref] [PubMed]
- Zou M, Zhang Q. miR-17-5p accelerates cervical cancer cells migration and invasion via the TIMP2/MMPs signaling cascade. Cytotechnology 2021;73:619-27. [Crossref] [PubMed]
- Kim TW, Lee YS, Yun NH, et al. MicroRNA-17-5p regulates EMT by targeting vimentin in colorectal cancer. Br J Cancer 2020;123:1123-30. [Crossref] [PubMed]
- Jin L, Luo Y, Zhao YC, et al. MiR-183-5p Promotes Tumor Progression of Osteosarcoma and Predicts Poor Prognosis in Patients. Cancer Manag Res 2021;13:805-14. [Crossref] [PubMed]
- Yang M, Liu R, Li X, et al. miRNA-183 suppresses apoptosis and promotes proliferation in esophageal cancer by targeting PDCD4. Mol Cells 2014;37:873-80. [Crossref] [PubMed]
- Bavelloni A, Ramazzotti G, Poli A, et al. MiRNA-210: A Current Overview. Anticancer Res 2017;37:6511-21. [PubMed]


