
Prostate cancer bioinformatics analysis: emerging genomic profiling techniques
Diagnosis techniques
Serum prostate-specific antigen (PSA) level measurement has been used as a diagnosis and prognostic measure for prostate cancer (PCa) (1,2). Catalona et al. reported that serum PSA measurement is a useful adjunct to rectal examination and ultrasonography in PCa diagnosis (1). Cooper et al. studied PSA levels in metastatic PCa cohorts of patients. They reported that 44 of 60 patients had a decrease of PSA to ≤10 ng/mL at period of 3 to 6 months after the EORTC trial of Zoladex plus flutamide versus orchidectomy (2). The European Association of Urology (EAU) suggested using a systematic prostate biopsy under ultrasound guidance and local anesthesia as a preferred diagnostic method than measuring PSA level (3). However, this method comes with underdetection and under-grading of clinically significant (4). ElKarami et al. applied a machine learning approach to magnetic resonance imaging-guided targeted biopsy (MRI-TB) on a cohort of patients who upgraded to significant PCa on MRI-TB was defined as upgrading to Gleason score (3+4) to Gleason score (4+3) (5). MRI-TB works with a visual diagnosis of lesions or the shape of the cell in the tissue (6).
Wei et al. introduced a bioinformatics pipeline that identified potential hub genes biomarkers for PCa diagnosis and prognosis. The methods were applied to four Gene Expression Omnibus (GEO) datasets that contain 123 PCa samples versus 76 normal. A panel of 368 differentially expressed genes (DEGs) was identified, including 120 up-regulated DEGs and 248 down-regulated DEGs. Pathway analysis showed that those DEGs were enriched in focal adhesion, chemical carcinogenesis, drug metabolism, and cytochrome P450 pathways. Then protein-protein interaction (PPI) analysis identified 11 hub genes network of the DEGs. While the work is comprehensive, DEG from microarray can be a starting point for biomarker identification (7). However, modern next-generation sequencing (NGS) allows deeper throughput into genomic insights. Hamzeh et al. proposed a machine learning approach to identify Gleason stages biomarkers based on NGS data. Genes transcripts from RNA-Seq data could determine a Gleason stage from the rest of the stages (8). The transcriptomics method identified biomarkers related to genes strongly associated with the progression of PCa, including PIAS3, UBE2V2, and EPB41L1.
The technical advancement in biomedical engineering technology allowed various measures from various omics areas, including genomics, transcriptomics, epigenomics, proteomics, metabolomics, and many other biomedical fields. The authors in (9) proposed a deep learning approach to predict the relapse in PCa. The multi-omics model integrates five different omics: mRNA, miRNA, DNA methylation, copy number variations (CNVs), and long non-coding RNA (lncRNA) by utilizing H2O package. Gholami et al. highlighted the importance of utilizing multi-omic approaches to improve outcomes in treating PCa patients. They survey recent works that applied a multi-omics data integration model to analyze PCa. The authors highlighted the challenges of using multi-omics approaches; it is an invasive biopsical practice to collect data with many side effects. The heterogenity nature of the various multi-omics data may result in a biased model. The resulting different molecular characteristics of tumor cells lead to effective screening methods for early cancer detection, patient selection strategies, or treatment response assessment (10).
Technology reflection
While microarray technology may provide relative gene expressions affordably, it comes with many drawbacks, including identifying only known transcripts with low sensitivity and providing no alternative splicing information. Conversely, RNA-Seq throughout NGS technology brought more high-intensity transcriptomics events and measurements. It can also identify unknown transcripts throughout de novo reads alignment technique (11). The NGS technology is becoming cheaper and expected to be affordable by the next couple of years for genomic profiling.
Similar to RNA-Seq library preparation, microarray requires converting RNA to cDNA. However, it requires an additional step, hybridization into microarray, before scanning, as seen in Figure 1. While RNA-Seq requires amplification before sequencing and extra data processing after sequencing, including reads alignments to the human genome and transcript assembly, as seen in Figure 1. Multi-omics data integration studies rely on integrating data from different omics measurements, which yields to comprehensive analysis of the disease that can extract various types of biomarkers. However, it is an expensive approach and complicated to analyze (10). Table 1 highlights the strengths and drawbacks for different techniques of studying PCa.
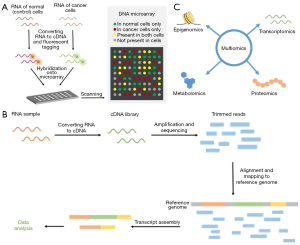
Table 1
| Properties/technology | Microarray | RNA-Seq | Multi-omics |
|---|---|---|---|
| Strengths | Low cost | High sensitivity | Comprehensive analysis |
| Well-known hybridization protocol | Transcription level biomarkers | Various types of biomarkers | |
| De novo assembly for unknown transcripts | |||
| Drawbacks | Low sensitivity | Costly | Very costly |
| Only gene level biomarkers | Preprocessing is required before data analysis | Each omic requires it’s own protocol | |
| It works for only known transcripts | Still no well-formulated integrative model | ||
| Low variance expression |
While microarray and RNA-Seq technology provide insight into gene expressions in the tumor tissue, the current trend of PCa bioinformatics analysis is to integrate different omics to find various biomarkers for the diagnosis and prognosis of the disease. With the power of artificial intelligent methods, the future direction is to integrate omics data with other types of medical data, including medical images including MRI, to predict the outcome of the PCa. The fusion of various health data may unveil the potential of the prohibition and treatment of the disease.
Acknowledgments
Funding: This work was partially funded by
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2423/coif). AA received financial support as a seed fund from the King Abdullah I School of Graduate Studies and Scientific Research at the Princess Sumaya University for Technology with grant number 2021/2022–25[16]. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Catalona WJ, Smith DS, Ratliff TL, et al. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med 1991;324:1156-61. [Crossref] [PubMed]
- Cooper EH, Armitage TG, Robinson MR, et al. Prostatic specific antigen and the prediction of prognosis in metastatic prostatic cancer. Cancer 1990;66:1025-8. [Crossref] [PubMed]
- Heidenreich A, Bastian PJ, Bellmunt J, et al. EAU guidelines on prostate cancer. part 1: screening, diagnosis, and local treatment with curative intent-update 2013. Eur Urol 2014;65:124-37. [Crossref] [PubMed]
- Schouten MG, van der Leest M, Pokorny M, et al. Why and Where do We Miss Significant Prostate Cancer with Multi-parametric Magnetic Resonance Imaging followed by Magnetic Resonance-guided and Transrectal Ultrasound-guided Biopsy in Biopsy-naïve Men? Eur Urol 2017;71:896-903. [Crossref] [PubMed]
- ElKarami B, Deebajah M, Polk S, et al. Machine learning-based prediction of upgrading on magnetic resonance imaging targeted biopsy in patients eligible for active surveillance. Urol Oncol 2022;40:191.e15-20. [Crossref] [PubMed]
- Drost FH, Osses DF, Nieboer D, et al. Prostate MRI, with or without MRI-targeted biopsy, and systematic biopsy for detecting prostate cancer. Cochrane Database Syst Rev 2019;4:CD012663. [Crossref] [PubMed]
- Wei T, Liang Y, Anderson C, et al. Identification of candidate hub genes correlated with the pathogenesis, diagnosis, and prognosis of prostate cancer by integrated bioinformatics analysis. Transl Cancer Res 2022;11:3548-71. [Crossref] [PubMed]
- Hamzeh O, Alkhateeb A, Rezaeian I, et al. Finding transcripts associated with prostate cancer gleason stages using next generation sequencing and machine learning techniques. In: International Conference on Bioinformatics and Biomedical Engineering. Cham: Springer, 2017.
- Wei Z, Han D, Zhang C, et al. Deep Learning-Based Multi-Omics Integration Robustly Predicts Relapse in Prostate Cancer. Front Oncol 2022;12:893424. [Crossref] [PubMed]
- Gholami N, Haghparast A, Alipourfard I, et al. Prostate cancer in omics era. Cancer Cell Int 2022;22:274. [Crossref] [PubMed]
- Schulz MH, Zerbino DR, Vingron M, et al. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012;28:1086-92. [Crossref] [PubMed]


