
The stool syndecan2 methylation test is more robust than blood tests for methylated septin9, CEA, CA19-9 and CA724: a diagnostic test for the early detection of colorectal neoplasms
Introduction
Although colorectal cancer (CRC) is the second leading cause of death due to cancer (1), decreased mortality has been observed in a large number of countries, especially high-income countries, over the last decade (2). This reduction can be primarily attributed to early detection efforts and improved treatments (3,4). Most CRCs develop from adenomas, with a very slow progression from benign adenomas to invasive carcinoma (5). As one of the most preventable cancers (6), CRC can be successfully treated if detected early; and screening substantially reduces the risk of CRC (5,7).
Currently, colonoscopy is the gold standard for the early diagnosis of CRC. However, due to its invasiveness, need for bowel preparation and complications, compliance in the public remains very low, which impedes screening efforts. A recent CRC screening program based on more than a million participants in China reported a colonoscopy participation rate of only 14.0% among high-risk individuals (8). Sigmoidoscopy or computed tomography (CT) colonography, other high-sensitivity screening methods, are semi-invasive tests that also have limitations (9). The fecal occult blood test (FOBT) and fecal immunochemical test (FIT) are the two most widely used noninvasive stool tests for CRC due to their convenience and relatively low costs (10). However, both the FOBT and FIT have relatively limited sensitivities (11-13). Thus, a noninvasive, highly accurate screening method to detect CRC at an early stage is urgently needed, especially for those who are reluctant to undergo colonoscopy examinations.
Aberrant DNA methylation is the most prevalent epigenetic alteration that occurs in all stages of carcinogenesis, and it can be successfully detected in several types of biological samples (blood, tissue, stool) (14,15). Due to their biological rationality and user-friendly nature, DNA methylation-based biomarkers are valuable tools in the early detection of CRC (16). To date, few studies have assessed the performance of stool methylated syndecan2 (mSDC2) analysis in detecting colorectal neoplasms (17-22), with a sensitivity ranging from 77.4% to 90.2% and a specificity ranging from 88.2% to 98.0%. However, results regarding the sensitivities of analyses performed according to tumor stages were inconsistent, with some showing a higher sensitivity for the detection of early stages I/II (20,21), while others showed contrary results (18,22); the performance of this analysis for the assessment of various tumor locations also remains unclear. In addition, it has been suggested that levels of SDC2 methylation are increased in other types of cancer tissues, such as in glioma and gastric cancer (23,24). However, few studies have explored the potential factors associated with the false-positive rate of stool SDC2 methylation tests.
Methylated septin9 (mSEPT9) is thought to be released from apoptotic cells shed from solid tumors into the plasma in CRC patients (25,26). Increased concentrations of circulating mSEPT9 in the blood have been reported to be positively correlated with tumor burden (27), yielding a sensitivity of 52% to 73% and a specificity of 84% to 91% for CRC detection (27-30). Whereas, the evidence on its performance in detecting precancerous lesions remains limited and unclear, given the histological definition of precancers that do not invade the basement membrane or underling blood vessels.
Serum tumor biomarkers, including carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9) and carbohydrate antigen 724 (CA724), have been widely used for the screening, diagnosis and surveillance of gastrointestinal (GI) cancer but have insufficient sensitivity and specificity (31-33). Thus, developing a more reliable method is urgently needed. To date, no previous studies have compared the performance of the stool SDC2 test with that of analyses of these serum biomarkers.
Thus, we aimed to evaluate and compare the clinical performance of a stool SDC2 methylation test and blood tests for mSEPT9, CEA, CA19-9 and CA724 in the identification of patients with CRC or adenoma. We present the following article in accordance with the STARD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1710/rc).
Methods
Study population
Stool samples were obtained from patients in Tianjin Union Medical Center between February 2021 and March 2022. The target population was enrolled, including patients with a definitive or suspected diagnosis of CRC, those carrying benign gastroenterological lesions such as hemorrhoids and polyps, those with other types of cancers, and those with no evidence of disease (NED). All stool samples were collected prior to colonoscopy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Tianjin Union Medicine Center (IRB number: 2021-B37). Written informed consent was obtained from all participants for obtaining blood or stool samples.
Stool sample collection
Every participant was required to provide a 1.5 to 10 g (average 4.5 g) stool sample in a semiquantitative stool collection device prefilled with 15 mL of preservative buffer (Creative Biosciences, Guangzhou, China). All buffered stools were immediately transported to our laboratory and homogenized and centrifuged subsequently. The supernatant was aliquoted and frozen at −80 ℃ for subsequent processing.
Stool SDC2 methylation test
A methylation-specific detection kit (Creative Biosciences) was used to qualitatively assess the methylation of the human SDC2 gene promoter region in the stool samples. The tests were performed strictly in accordance with the manufacturer’s protocol, which has been previously described in detail (17). Briefly, the test consisted of two steps: DNA extraction and transformation and real-time quantitative methylation-specific polymerase chain reaction (PCR) (qMSP). The SDC2 gene and β-actin (ACTB) gene were simultaneously captured by a magnetic bead. Stool DNA was purified and enriched using sequence-specific capture technology as reported previously with minor modifications (34). The unmethylated DNA was then transformed with sulfites, whereas the methylated SDC2 genes were not. qMSP was performed to quantitatively detect SDC2 and ACTB methylation status in stool samples. PCR amplification was performed under the following cycling conditions: 95 ℃ for 5 min, 48 cycles at 95 ℃ for 20 s, 58 ℃ for 60 s, and 72 ℃ for 30 s, and a final cooling step at 37 ℃ for 30 s. All valid samples satisfied the required cycle threshold (Ct) value of ACTB levels less than or equal to 36. Positivity for SDC2 methylation was defined as a Ct value less than or equal to 38, while negativity for SDC2 methylation was defined as a Ct value greater than 38 or undetected Ct values.
Blood tests for mSEPT9, CEA, CA19-9 and CA724 levels
All these tests were performed in the Clinical Laboratory of Tianjin Union Medical Center. The blood-based Septin9 gene methylation assay was performed with an improved SEPT9 kit (Epigenomics AG for Epi proColon 2.0) following the provided instructions (35). The levels of serum tumor biomarkers CEA, CA19-9 and CA724 were measured by electrochemiluminescence. Positive values were defined by widely accepted cutoffs (CEA: 5 ng/mL, CA19-9: 37 U/mL, CA242: 20 U/mL). Positivity for SETP9 methylation was defined as a Ct value less than or equal to 41, while negativity for mSEPT9 methylation was defined as a Ct value greater than 41 or undetected Ct values.
Colonoscopy examinations
The gold diagnosis was colonoscopy combined with pathology report of tissue biopsy. All endoscopic examinations were performed in our medical center by experienced endoscopists who had at least 5 years of experience and were all board certified to perform endoscopy. All abnormal findings were confirmed by expert gastrointestinal (GI) pathologists following up-to-date clinical guidelines. Data from only high-quality colonoscopies were included, with adequate bowel preparation, photo documentation of cecal landmarks, and a withdrawal time greater than 6 min. Tumor-node-metastasis (TNM) stage was defined according to the guidelines of the 7th edition of the Cancer Staging Manual from the American Joint Committee on Cancer. To evaluate the performance of mSDC2 analysis in detecting colorectal neoplasms, colonoscopy findings were categorized into 3 groups: CRC, adenoma and normal colonoscopy, with the latter referring to a colonoscopy during which no adenoma or CRC was found. When a participant was diagnosed with two or more types of colorectal lesions, only the most advanced lesion was used for classification.
Statistical analysis
If mSEPT9 and mSDC2 levels were not detected, the Ct value of mSEPT9 levels was set to 42.0 (the maximal number of PCR cycles for mSEPT9), and the Ct value of mSDC2 levels was set to 48.0 (the maximal number of PCR cycles for mSDC2).
The chi-square test was employed to compare the qualitative methylation levels and clinicopathological features among patients. Sensitivity (equal to positive detection rate of CRC or adenoma) is defined as true positives/(true positives + false negatives), which implies the probability to identify diseased persons correctly using colonoscopy combined with pathology report as gold standard. Likewise, the specificity is defined as true negatives/(true negatives + false positives), which implies the probability to identify non-diseased person correctly. McNemar’s chi-squared test with continuity correction was performed to compare sensitivity and specificity between the two methods. The Wilcoxon test was performed to compare levels between two testing subjects. Receiver operating characteristic (ROC) curves and the area under the curve (AUC) were used to assess predictive performance, and DeLong’s test was performed to compare two correlated ROC curves. A multivariable logistic regression model was performed to further explore clinicopathological features associated with stool SDC2 methylation, and odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. All analyses were performed using R software (V.4.1.2). Results with two-sided P values less than 0.05 were considered statistically significant.
Results
Patients and lesion characteristics
A total of 1,002 eligible patients with valid stool SDC2 methylation tests prior to colonoscopy were included in the present study, among whom 457 (45.6%) were tested for plasma mSEPT9, 601 (60.0%) were tested for serum CEA, 516 (51.5%) were tested for serum CA19-9, and 507 (50.6%) were tested for serum CA724. The demographic and lesion characteristics are shown in Table 1. There were a total of 445 (44.4%) CRC patients, including 24 patients with stage 0, 51 with stage I, 151 with stage II, 164 with stage III, 29 with stage IV and 26 patients with an unknown stage. In addition, 115 (11.5%) patients were diagnosed with adenoma, 357 (35.6%) patients with nonneoplastic GI diseases (including 34 polyps, 224 hemorrhoids, 55 perianal fistulas, 14 abscesses and 30 other diagnoses), 16 (1.6%) patients with non-CRC cancers (including 6 GI cancers and 10 non-GI cancers), 7 (0.7%) patients with other non-GI disorders, and 62 (6.2%) patients with NED.
Table 1
| Characteristic | Patients tested with stool mSDC2 (n=1,002) | Patients tested with plasma mSEPT9 (n=457) | Patients tested with CEA (n=601) | Patients tested with CA19-9 (n=516) | Patients tested with CA724 (n=507) |
|---|---|---|---|---|---|
| Age, n (%) | |||||
| <60 years | 455 (45.4) | 149 (32.6) | 195 (32.4) | 164 (31.8) | 164 (32.3) |
| ≥60 years | 547 (54.6) | 308 (67.4) | 406 (67.6) | 352 (68.2) | 343 (67.7) |
| Sex, n (%) | |||||
| Male | 630 (62.9) | 298 (65.2) | 391 (65.1) | 332 (64.3) | 323 (63.7) |
| Female | 372 (37.1) | 159 (34.8) | 210 (34.9) | 184 (35.7) | 184 (36.3) |
| Lesion, n (%) | |||||
| CRC | 445 (44.4) | 340 (74.4) | 414 (68.9) | 352 (68.2) | 338 (66.7) |
| TNM stage | |||||
| 0 | 24 (5.4) | 20 (5.9) | 23 (5.6) | 20 (5.7) | 20 (5.9) |
| I | 51 (11.5) | 35 (10.3) | 44 (10.6) | 37 (10.5) | 37 (10.9) |
| II | 151 (33.9) | 115 (33.8) | 139 (33.6) | 117 (33.2) | 112 (33.1) |
| III | 164 (36.9) | 125 (36.8) | 154 (37.2) | 132 (37.5) | 125 (37.0) |
| IV | 29 (6.5) | 23 (6.8) | 29 (7.0) | 25 (7.1) | 23 (6.8) |
| Unknown | 26 (5.8) | 22 (6.5) | 25 (6.0) | 21 (6.0) | 21 (6.2) |
| Location† | |||||
| Right colon | 55 (12.4) | 46 (13.5) | 50 (12.1) | 43 (12.2) | 39 (11.5) |
| Left colon | 151 (33.9) | 111 (32.6) | 140 (33.8) | 112 (31.8) | 111 (32.8) |
| Rectum | 231 (51.9) | 177 (52.1) | 217 (52.4) | 190 (54.0) | 181 (53.6) |
| Unspecific | 8 (1.8) | 6 (1.8) | 7 (1.7) | 7 (2.0) | 7 (2.1) |
| Differentiation | |||||
| Well | 72 (16.2) | 55 (16.2) | 68 (16.4) | 64 (18.2) | 58 (17.2) |
| Moderate | 243 (54.6) | 187 (55.0) | 222 (53.6) | 184 (52.3) | 178 (52.7) |
| Poor | 39 (8.8) | 29 (8.5) | 36 (8.7) | 30 (8.5) | 29 (8.6) |
| Unknown | 91 (20.4) | 69 (20.3) | 88 (21.3) | 74 (21.0) | 73 (21.6) |
| Adenoma, n (%) | 115 (11.5) | 30 (6.6) | 45 (7.5) | 43 (8.3) | 42 (8.3) |
| Non-neoplastic GI diseases, n (%) | 357 (35.6) | 31 (6.8) | 60 (10.0) | 48 (9.3) | 50 (9.9) |
| Non-CRC cancers, n (%) | 16 (1.6) | 12 (2.6) | 16 (2.7) | 11 (2.1) | 13 (2.6) |
| Other non-GI disorders, n (%) | 7 (0.7) | 4 (0.9) | 6 (1.0) | 4 (0.8) | 4 (0.8) |
| NED, n (%) | 62 (6.2) | 40 (8.8) | 60 (10.0) | 58 (11.2) | 60 (11.8) |
†, right colon was defined as proximal to splenic flexure, while left colon was defined as from splenic flexure to the rectosigmoid junction. mSDC2, methylated syndecan2; mSEPT9, methylated septin9; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9; CA724, carbohydrate antigen 724; CRC, colorectal cancer; TNM, tumor-node-metastasis; GI, gastrointestinal; NED, no evidence of disease.
Notably, as the detection of CRC was the primary goal, patients who had adenoma, nonneoplastic GI diseases, non-CRC cancers, and NEDs were all grouped together and treated as control cases in the analyses. For the detection of adenoma, patients with nonneoplastic GI diseases, non-CRC cancers, and NED were combined and classified as “controls”.
Comparison of clinical performance in detecting neoplasms
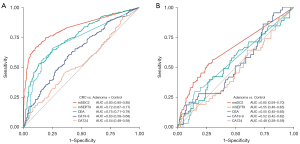
The AUC for stool mSDC2 test in detecting CRC among all subjects was 0.83 (95% CI: 0.80–0.85), whereas for testing mSEPT9, CEA, CA19-9 and CA724 in blood were 0.72 (95% CI: 0.67–0.77), 0.75 (95% CI: 0.71–0.79), 0.63 (95% CI: 0.58–0.68) and 0.54 (95% CI: 0.48–0.59), respectively (Figure 1A). DeLong’s test showed that the differences between stool mSDC2 and all other biomarkers exhibited P values that were less than 0.05. In the detection of adenoma, the AUC for mSDC2 was 0.65 (95% CI: 0.59–0.70), which was greater than that for mSEPT9 (AUC: 0.55; 95% CI: 0.46–0.65; P=0.014) and CA724 (AUC: 0.48; 95% CI: 0.38–0.58; P=0.01) but was not different from that for CEA (AUC: 0.55; 95% CI: 0.45–0.65; P=0.072) and CA19-9 (AUC: 0.52; 95% CI: 0.42–0.62; P=0.08) (Figure 1B).
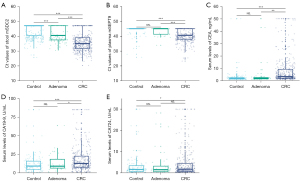
The levels of mSDC2 in stool and mSEPT9 in plasma and serum levels of CEA, CA19-9, and CA724 in the CRC, adenoma and control groups are shown in Figure 2. The levels of all five markers were significantly higher in CRC patients than in the control group, but only stool mSDC2 could be used significantly to distinguish adenoma patients from controls.
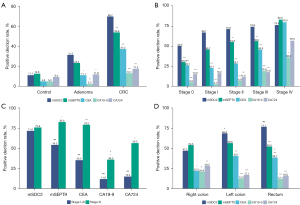
The positive detection rates in the CRC, adenoma and control groups are shown in Figure 3A. The stool mSDC2 test detected 69.7% of CRCs, which was a significantly higher rate than that detected by mSEPT9, CEA, CA19-9 and CA724 (53.8%, 37.2%, 13.1% and 17.5%, respectively, all P<0.001). In the detection of adenoma, the detection rate of the stool mSDC2 test was 31.3%, which was significantly higher than that of CEA (11.1%, P=0.022), CA19-9 (2.3%, P=0.002) and CA724 (11.9%, P=0.027) and higher than the 23.3% of plasma mSEPT9, although the difference was not significant.
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of these five biomarkers in detecting CRC and adenomas are shown in Table 2. The NPVs of mSDC2, mSEPT9, CEA, CA19-9 and CA724 for the detection of CRC were 78%, 39%, 40%, 34% and 35%, respectively, with accuracies of 78%, 62%, 55%, 39% and 42%, respectively.
Table 2
| Test | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | Accuracy (95% CI) |
|---|---|---|---|---|---|
| Detection of CRC | |||||
| mSDC2 | 70% (65–74%) | 85% (81–88%) | 78% (74–82%) | 78% (74–81%) | 78% (75–80%) |
| mSEPT9 | 54% (48–59%) | 85% (77–91%) | 91% (86–95%) | 39% (33–45%) | 62% (57–66%) |
| CEA | 37% (33–42%) | 94% (89–97%) | 93% (88–96%) | 40% (36–45%) | 55% (51–59%) |
| CA19-9 | 13% (10–17%) | 96% (91–98%) | 87% (75–95%) | 34% (30–38%) | 39% (35–44%) |
| CA724 | 17% (14–22%) | 90% (84–94%) | 78% (67–86%) | 35% (31–40%) | 42% (37–46%) |
| Detection of adenoma | |||||
| mSDC2 | 31% (23–41%) | 89% (86–92%) | 42% (32–54%) | 83% (80–87%) | 77% (73–80%) |
| mSEPT9 | 23% (10–42%) | 87% (79–94%) | 39% (17–64%) | 77% (67–85%) | 71% (62–79%) |
| CEA | 11% (4–24%) | 95% (90–98%) | 42% (15–72%) | 77% (70–83%) | 75% (68–81%) |
| CA19-9 | 2% (0–12%) | 95% (90–98%) | 14% (0–58%) | 73% (66–80%) | 71% (63–78%) |
| CA724 | 12% (4–26%) | 91% (84–95%) | 29% (10–56%) | 76% (68–82%) | 71% (64–78%) |
mSDC2, methylated syndecan2; mSEPT9, methylated septin9; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9; CA724, carbohydrate antigen 724; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; CRC, colorectal cancer.
Rates of the detection of various CRC stages
Among the 419 patients staged based on the surgically resected specimens, the respective detection rates by stool mSDC2 for patients with stage 0, I, II, III, and IV CRC were 50.0%, 66.7%, 70.9%, 73.8% and 75.9%, respectively, and did not differ significantly among stages (P=0.166). Regarding the stage-specific sensitivities, stool mSDC2 performed better than other biomarkers except for stage IV, followed by plasma mSEPT9 (Figure 3B). When patients with stages I–III cancer were combined, stool mSDC2 detected 71.6% of CRCs, which was significantly higher than 54.2% by mSEPT9, 35.3% by CEA, 11.9% by CA19-9 and 15.0% by CA724 (all P<0.001). However, detection rates for stage IV CRC did not differ significantly between mSDC2 and mSEPT9, CEA or CA724 (82.6%, 75.9%, 79.3% and 56.5%, respectively, all P>0.05), with mSDC2 exhibiting an advantage only over CA19-9 (36.0%, P=0.016). Interestingly, mSEPT9 and CEA showed even higher detection rates than mSDC2 for stage IV CRC.
All tests except for stool mSDC2 had significantly higher sensitivities for detecting stage IV CRC than for detecting stage I–III CRC (Figure 3C). However, stool mSDC2 could distinguish patients with stage I–III CRC from those with stage 0 CRC well, while other biomarkers could not. The levels of mSDC2 in stool, mSEPT9 in plasma, and CEA, CA19-9, and CA724 in serum stratified according to CRC stage are shown in Figure S1. Similar to the sensitivity results, the methylation level of SDC2 increased with stage but was not higher in stage IV CRC, while the levels of biomarkers assessed in other tests were significantly higher in patients with stage IV cancer than in patients at any other stage.
Detection rates in various locations
A multivariable logistic regression model was created to assess the association between stool SDC2 methylation and clinicopathological features (Table 3). After adjusting for age (<60 and ≥60 years), sex, tumor stage, location, and differentiation, we found that only location was significantly associated with a positive result from the stool mSDC2 test. With the proximal colon as the reference, the ORs were 4.54 for detecting cancer in the distal colon (P<0.001) and 6.26 for detecting cancer in the rectum (P<0.001). Sex, age, stage and differentiation had no effect on positive or negative test results (all P>0.05).
Table 3
| Feature | Number (%) | Crude OR (95% CI) | Adjusted OR (95% CI) | P value |
|---|---|---|---|---|
| Age | ||||
| <60 years | 117 (26.3) | Reference | Reference | |
| ≥60 years | 328 (73.7) | 0.92 (0.54–1.58) | 0.75 (0.42–1.36) | 0.345 |
| Sex | ||||
| Male | 295 (66.3) | Reference | Reference | |
| Female | 150 (33.7) | 1.33 (0.80–2.20) | 1.54 (0.88–2.68) | 0.127 |
| Location | ||||
| Right colon | 55 (12.6) | Reference | Reference | |
| Left colon | 151 (34.6) | 4.36 (2.12–8.95) | 4.54 (2.12–9.75) | <0.001 |
| Rectum | 231 (52.9) | 5.72 (2.85–11.48) | 6.26 (2.99–13.1) | <0.001 |
| Differentiation | ||||
| Well | 72 (20.3) | Reference | Reference | |
| Moderate | 243 (68.6) | 0.69 (0.36–1.30) | 0.64 (0.32–1.27) | 0.201 |
| Poor | 39 (11.0) | 0.46 (0.19–1.10) | 0.54 (0.20–1.47) | 0.229 |
| Stage | ||||
| 0 | 24 (5.7) | Reference | Reference | |
| I | 51 (12.2) | 4.57 (0.38–54.66) | 1.49 (0.09–23.76) | 0.777 |
| II | 151 (36.0) | 5.25 (0.46–59.51) | 2.35 (0.16–34.04) | 0.531 |
| III | 164 (39.1) | 5.49 (0.48–62.22) | 2.07 (0.14–30.18) | 0.595 |
| IV | 29 (6.9) | 14.00 (0.58–338.78) | 4.05 (0.14–117.86) | 0.416 |
CRC, colorectal cancer; OR, odds ratio; CI, confidence interval.
The positive detection rates according to tumor location are shown in Figure 3D. The sensitivities did not vary significantly between CRCs in the rectum and distal colon (77.1% vs. 68.9%, P>0.05), but both had significantly higher sensitivities than the proximal colon (77.1% vs. 47.2%, P<0.001; 68.9% vs. 47.2%, P=0.007). CEA showed a similar trend to SDC2, and the sensitivities of the other biomarkers were not significantly influenced by location.
False-positive rates
The positive detection rates of mSDC2, mSEPT9, CEA, CA19-9 and CA724 in the control group were 11.1%, 12.6%, 4.9%, 5.3% and 9.4%, respectively, with no significant variation. The positive rates of mSDC2 and mSEPT9 were high, which may have been partly due to the presence of interfering diseases. Details regarding the types of interfering diseases and the outcomes of the tests are shown in Table S1. Stool mSDC2 detected 6/16 (37.5%) non-CRC cancers (including one small intestine cancer, one prostate cancer, three lymphomas and one lung cancer) and 42/357 (11.8%) nonneoplastic GI diseases. Plasma mSEPT9 detected 4/12 (33.3%) non-CRC cancers (including three prostate cancers and one lung cancer) and 4/31 (12.9%) nonneoplastic GI diseases.
Discussion
In a clinical setting, earlier detection of CRC is still the most effective method to reduce morbidity and mortality (1,36). A high-risk factor questionnaire plus the FOBT is currently the preliminary CRC screening strategy in China, but accumulated data showed a low PPV for selecting high-risk individuals (37,38). Moreover, even when identified as high-risk individuals, only 14.0% underwent a colonoscopy as recommended (8). Thus, a noninvasive screening method with high sensitivity in detecting early-stage CRC and precancerous lesions is urgently needed. In this comparison of testing methods using inpatient cases, we found that both nonmetastatic CRCs and colorectal adenomas were detected at significantly higher rates by the stool SDC2 methylation test than by blood tests for mSEPT9, CEA, CA19-9 or CA724.
Although both methylated SDC2 and SEPT9 are broadly expressed in adenoma tissue (17,27,28), the detection rate by plasma mSEPT9 remained low. In the present study, the detection rate for adenoma by plasma mSEPT9 was only 23.3%, which was comparable to the 14–18% reported by previous studies (28,30,39,40). Moreover, plasma mSEPT9 detected significantly fewer early-stage CRC patients than stool mSDC2; however, for stage IV CRC, mSEPT9 had a higher detection rate than stool mSDC2, although the difference was not significant.
This result can be explained by the key differences in how the markers are released into the blood and stool (40). While methylated SDC2 is released into the stool through luminal exfoliation, circulating methylated SEPT9 is released into plasma from apoptotic cells that are shed from the tumor (25,26), with the latter appearing to be associated with vascular invasion. Thus, based on the histological definition, which is that there is no invasion of the basement membrane or underling blood vessels in adenomas, there is no route for mSEPT9 entry into the blood. Moreover, exfoliation into stool generally occurs earlier than vascular invasion into blood during the progression of CRC, limiting plasma DNA testing for early-stage CRC (40). This explains why plasma mSEPT9 had higher sensitivity for detecting patients with distant metastasis than for those with early-stage CRCs (39,41). Therefore, plasma mSEPT9 is more suitable for monitoring recurrence than for early screening.
While the levels of mSEPT9, CEA, CA19-9 and CA724 in blood tended to be highest in patients with stage IV disease, there appeared to be an opposite trend for mSDC2 levels in stool. A similar trend was also reported by two previous studies targeting other methylated genes in stool (BMP3, NDRG4, vimentin, and TFPI2), in which the detection rates for stage IV CRC were significantly lower than those for stage I–III CRC (40,42). This finding might be explained by the fact that DNA hypomethylation occurs with late progression (43), surface necrosis occurs with reduced colonocyte exfoliation, or additional factors. Even so, prior studies focusing on mSDC2 as well as our study suggest that this marker has a superior sensitivity for more advanced stages, although the differences were not significant (18,20,22).
Although the FOBT is widely used in CRC screening, its sensitivity for the early screening of CRC is limited. With intermittent occult bleeding, repeated FOBTs are needed to guarantee its efficacy (44,45). In addition, unlike the FOBT, the stool mSDC2 test requires no diet or medication restrictions that might result in false-positive or false-negative outcomes (46), except for berberine, a Chinese herbal medicine (19).
It has been suggested that the SDC2 methylation level is elevated in some types of cancer tissues, such as in glioma and gastric cancer (23,24). However, it remains unclear whether interfering diseases influence the accuracy of stool SDC2 methylation tests in detecting CRC. A multicenter clinical study by Wang et al. reported that stool mSDC2 analysis could not be used to detect any of the cancers in the digestive tract (0/30) (20). Cooper et al. reassessed false-positive stool DNA tests in 30 patients and did not find non-colorectal lesions by repeat colonoscopy, upper endoscopy or a review of the medical records 11–29 months after the initial test (47). However, in another clinical trial by Han et al., 30.4% of gastric cancer patients (7/23) and 30% of liver cancer patients (3/10) tested positive for SDC2 methylation in the stool (18). In our study population, tests for SDC2 methylation were positive in 37.5% (6/16) of patients with other types of cancers; for those with other nonneoplastic GI diseases, the detection rate was 11.8% (42/357). However, Wang et al. reported only 1 positive mSDC2 test result among 21 patients (4.7%) with digestive tract ailments (20). This difference might have resulted from the distribution of diseases, with 90% of the patients in their study exhibiting nonneoplastic GI diseases located in the upper digestive tracts, but over 80% of patients had anal and rectal diseases in our study. Hence, further research is needed to define the effects of interfering diseases on SDC2 methylation in the stool.
Evidence suggests that plasma mSEPT9 is a potential biomarker for several types of cancer (48-50). In our study, 33.3% of non-CRC cancer patients and 12.9% of patients with nonneoplastic GI disease exhibited positivity for plasma mSEPT9 expression, which was consistent with other reports, with a rate of 30.8% (4/13) observed among patients with non-CRC cancers and a rate of 18.2% (2/11) observed among patients with non-CRC GI diseases (39,40). Given the high cost of the test and the presence of multiple potential contributors to elevated plasma levels, mSEPT9 analysis might be unsuitable for CRC screening.
This study had some limitations. First, this was a single-center study in only Asian patients. Second, the stool test for SDC2 methylation was performed on a hospital-based cohort with many interfering diseases; thus, the results may not be representative of the findings observed in asymptomatic individuals undergoing general screening. However, the test would be more effective if such differences hold, as this method is applied in a general screening setting. Third, the size of the adenomas and pathological information regarding villous and serrated adenomas were not collected, which further limited our analysis of their influence on the performance of the test.
Conclusions
In conclusion, this is the first study to compare a stool SDC2 methylation test with blood tests for mSEPT9, CEA, CA19-9 and CA724. We demonstrated that the stool SDC2 methylation test had a better performance for detecting adenoma and nonmetastatic CRC than the other biomarkers, confirming the clinical potential of this test as a new useful noninvasive screening tool for early-stage CRC. Further large-scale studies are needed to corroborate and expand upon these findings.
Acknowledgments
Funding: This study was supported by the National Key R&D Program of China (2017YFC1700606, 2017YFC1700604); Key R&D Projects in the Tianjin Science and Technology Pillar Program (19YFZCSY00420); Natural Science Foundation of Tianjin, Grant Number (21JCZDJC00060, 21JCYBJC00180); Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-044A).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1710/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1710/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1710/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66:683-91. [Crossref] [PubMed]
- Edwards BK, Ward E, Kohler BA, et al. Annual report to the nation on the status of cancer, 1975-2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer 2010;116:544-73. [Crossref] [PubMed]
- Meester RG, Doubeni CA, Lansdorp-Vogelaar I, et al. Colorectal cancer deaths attributable to nonuse of screening in the United States. Ann Epidemiol 2015;25:208-13.e1. [Crossref] [PubMed]
- Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988;319:525-32. [Crossref] [PubMed]
- Mannucci A, Zuppardo RA, Rosati R, et al. Colorectal cancer screening from 45 years of age: Thesis, antithesis and synthesis. World J Gastroenterol 2019;25:2565-80.
- Grady WM, Markowitz SD. The molecular pathogenesis of colorectal cancer and its potential application to colorectal cancer screening. Dig Dis Sci 2015;60:762-72. [Crossref] [PubMed]
- Chen H, Li N, Ren J, et al. Participation and yield of a population-based colorectal cancer screening programme in China. Gut 2019;68:1450-7. [Crossref] [PubMed]
- Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging 2016;11:967-76. [Crossref] [PubMed]
- Cross AJ, Wooldrage K, Robbins EC, et al. Faecal immunochemical tests (FIT) versus colonoscopy for surveillance after screening and polypectomy: a diagnostic accuracy and cost-effectiveness study. Gut 2019;68:1642-52. [Crossref] [PubMed]
- Lee JK, Liles EG, Bent S, et al. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med 2014;160:171. [Crossref] [PubMed]
- Ramdzan AR, Abd Rahim MA, Mohamad Zaki A, et al. Diagnostic Accuracy of FOBT and Colorectal Cancer Genetic Testing: A Systematic Review & Meta-Analysis. Ann Glob Health 2019;85:70. [Crossref] [PubMed]
- Tinmouth J, Lansdorp-Vogelaar I, Allison JE. Faecal immunochemical tests versus guaiac faecal occult blood tests: what clinicians and colorectal cancer screening programme organisers need to know. Gut 2015;64:1327-37. [Crossref] [PubMed]
- Berger BM, Ahlquist DA. Stool DNA screening for colorectal neoplasia: biological and technical basis for high detection rates. Pathology 2012;44:80-8. [Crossref] [PubMed]
- Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015;149:1204-25.e12. [Crossref] [PubMed]
- Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology 2010;138:2127-39. [Crossref] [PubMed]
- Niu F, Wen J, Fu X, et al. Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol Biomarkers Prev 2017;26:1411-9. [Crossref] [PubMed]
- Han YD, Oh TJ, Chung TH, et al. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin Epigenetics 2019;11:51. [Crossref] [PubMed]
- Su WC, Kao WY, Chang TK, et al. Stool DNA test targeting methylated syndecan-2 (SDC2) as a noninvasive screening method for colorectal cancer. Biosci Rep 2021;41:BSR20201930. [Crossref] [PubMed]
- Wang J, Liu S, Wang H, et al. Robust performance of a novel stool DNA test of methylated SDC2 for colorectal cancer detection: a multicenter clinical study. Clin Epigenetics 2020;12:162. [Crossref] [PubMed]
- Zhao G, Liu X, Liu Y, et al. Aberrant DNA Methylation of SEPT9 and SDC2 in Stool Specimens as an Integrated Biomarker for Colorectal Cancer Early Detection. Front Genet 2020;11:643. [Crossref] [PubMed]
- Oh TJ, Oh HI, Seo YY, et al. Feasibility of quantifying SDC2 methylation in stool DNA for early detection of colorectal cancer. Clin Epigenetics 2017;9:126. [Crossref] [PubMed]
- Foltz G, Yoon JG, Lee H, et al. DNA methyltransferase-mediated transcriptional silencing in malignant glioma: a combined whole-genome microarray and promoter array analysis. Oncogene 2009;28:2667-77. [Crossref] [PubMed]
- Chong Y, Mia-Jan K, Ryu H, et al. DNA methylation status of a distinctively different subset of genes is associated with each histologic Lauren classification subtype in early gastric carcinogenesis. Oncol Rep 2014;31:2535-44. [Crossref] [PubMed]
- Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 2010;38:6159-75. [Crossref] [PubMed]
- Mouliere F, El Messaoudi S, Pang D, et al. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol 2014;8:927-41. [Crossref] [PubMed]
- Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414-23. [Crossref] [PubMed]
- Tänzer M, Balluff B, Distler J, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One 2010;5:e9061. [Crossref] [PubMed]
- deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 2009;55:1337-46. [Crossref] [PubMed]
- Grützmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One 2008;3:e3759. [Crossref] [PubMed]
- Gao Y, Wang J, Zhou Y, et al. Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer. Sci Rep 2018;8:2732. [Crossref] [PubMed]
- Nicholson BD, Shinkins B, Pathiraja I, et al. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst Rev 2015;2015:CD011134. [Crossref] [PubMed]
- Shah R, Jones E, Vidart V, et al. Biomarkers for early detection of colorectal cancer and polyps: systematic review. Cancer Epidemiol Biomarkers Prev 2014;23:1712-28. [Crossref] [PubMed]
- Zou H, Taylor WR, Harrington JJ, et al. High detection rates of colorectal neoplasia by stool DNA testing with a novel digital melt curve assay. Gastroenterology 2009;136:459-70. [Crossref] [PubMed]
- Wu D, Zhou G, Jin P, et al. Detection of Colorectal Cancer Using a Simplified SEPT9 Gene Methylation Assay Is a Reliable Method for Opportunistic Screening. J Mol Diagn 2016;18:535-45. [Crossref] [PubMed]
- Kolligs FT. Diagnostics and Epidemiology of Colorectal Cancer. Visc Med 2016;32:158-64. [Crossref] [PubMed]
- Cai SR, Zhang SZ, Zhu HH, et al. Performance of a colorectal cancer screening protocol in an economically and medically underserved population. Cancer Prev Res (Phila) 2011;4:1572-9. [Crossref] [PubMed]
- Li JB, Qiu ZY, Deng YX, et al. Factors associated with positive predictive value of preliminary screening in a two-step screening strategy for colorectal neoplasms in China. Discov Oncol 2022;13:4. [Crossref] [PubMed]
- Sun J, Fei F, Zhang M, et al. The role of (m)SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer 2019;19:450. [Crossref] [PubMed]
- Ahlquist DA, Taylor WR, Mahoney DW, et al. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol 2012;10:272-7.e1. [Crossref] [PubMed]
- Xie L, Jiang X, Li Q, et al. Diagnostic Value of Methylated Septin9 for Colorectal Cancer Detection. Front Oncol 2018;8:247. [Crossref] [PubMed]
- Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 2012;142:248-56; quiz e25-6. [Crossref] [PubMed]
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene 2002;21:5400-13. [Crossref] [PubMed]
- Ahlquist DA, McGill DB, Fleming JL, et al. Patterns of occult bleeding in asymptomatic colorectal cancer. Cancer 1989;63:1826-30. [Crossref] [PubMed]
- Quintero E, Carrillo M, Gimeno-García AZ, et al. Equivalency of fecal immunochemical tests and colonoscopy in familial colorectal cancer screening. Gastroenterology 2014;147:1021-30.e1; quiz e16-7. [Crossref] [PubMed]
- Ransohoff DF, Lang CA. Screening for colorectal cancer with the fecal occult blood test: a background paper. American College of Physicians. Ann Intern Med 1997;126:811-22. [Crossref] [PubMed]
- Cooper GS, Markowitz SD, Chen Z, et al. Evaluation of Patients with an Apparent False Positive Stool DNA Test: The Role of Repeat Stool DNA Testing. Dig Dis Sci 2018;63:1449-53. [Crossref] [PubMed]
- Song L, Chen Y, Gong Y, et al. Opportunistic screening and survival prediction of digestive cancers by the combination of blood mSEPT9 with protein markers. Ther Adv Med Oncol 2020;12:1758835920962966. [Crossref] [PubMed]
- Jiao X, Zhang S, Jiao J, et al. Promoter methylation of SEPT9 as a potential biomarker for early detection of cervical cancer and its overexpression predicts radioresistance. Clin Epigenetics 2019;11:120. [Crossref] [PubMed]
- Constâncio V, Nunes SP, Moreira-Barbosa C, et al. Early detection of the major male cancer types in blood-based liquid biopsies using a DNA methylation panel. Clin Epigenetics 2019;11:175. [Crossref] [PubMed]


