
Clusterin is a biomarker of breast cancer prognosis and correlated with immune microenvironment
Highlight box
Key findings
• Clusterin is connected to prognosis of breast cancer patients and tumor immune cell infiltration.
What is known and what is new?
• Clusterin is involved in the invasion of immune cells in the tumor microenvironment.
• Enhanced clusterin expression was markedly associated with molecular typing of breast cancer and expression of multiple markers related to specific immune cell subsets.
What is the implication, and what should change now?
• Clusterin may be a biomarker of immune cell recruitment into breast tumors and an important biomarker for immune cell infiltration; consequently, being a valuable prognostic factor in breast cancer patients.
Introduction
Breast cancer is one of the most common diseases and main causes of women’s mortality worldwide (1). Breast tumors are heterogeneous, with five major subtypes (luminal A and B, HER2, basal, and normal), with different clinical characteristics and prognoses (2). In recent years, immune checkpoint inhibitors (ICIs) have been used in the treatment of malignant tumors. ICIs work by utilizing the host immune system to kill tumor cells (3). Currently, programmed death ligand-1 (PD-L1) is the only convincing predictive biomarker for ICIs in breast cancer, and clinical trials have concentrated on triple-negative breast cancer (4-7). Tumor-infiltrating immune cells are associated with prognosis, particularly tumor-associated macrophages (TAMs) and neutrophils, which are also associated with tumor chemotherapy (8). Therefore, clarifying the immune phenotype in the breast cancer microenvironment and how immune cells modulate breast cancer is essential to identify potential new immunotherapeutic targets.
Clusterin (CLU) is an evolutionarily conserved molecular chaperone present in diverse human tissues and fluids, and is considered an important tumor regulator (9-11). CLU regulates several cancer-related cellular events, including cancer cell proliferation, metastasis, stemness, epithelial-mesenchymal transition, survival, therapeutic resistance, and suppression of programmed cell death to support tumor growth and recurrence (12-14). It seems to vary its location and function to preserve cells and ensure their survival, and it is important in neuroprotection and tumors as well as in chemoresistance (15).
In the mouse splenic matrix, CLU mRNA is significantly downregulated after deletion of lymphoid receptors critical for development, maintenance, and function of secondary lymphoid organs (16). This is an early understanding of the function of CLU in the immune system. Semen CLU interaction with dendritic cell (DC)-specific intercellular-adhesion-molecule-captured non-integrins is reported to promote antigen capture by DCs and differentiation of DCs into tolerogens, characterized by an increased ability to promote expansion of Foxp3+ T regulatory (Treg) cells (17). Elevated preoperative secretory CLU expression in breast cancer correlates with cancer-associated fibroblast (CAF) resistance and tumor necrosis factor α (TNF-α)-induced apoptosis in breast cancer cells (18,19). Therefore, we speculated that CLU could be associated with tumor-infiltrating immune cells and affect the treatment of cancer patients.
Here, we performed a comprehensive assessment of the relationship between CLU and patient prognosis using multiple databases (PrognoScan, GEPIA, and Kaplan-Meier plotter), and explored the link between CLU and tumor immune cell infiltration using the Tumor Immunoassay Resource (TIMER). Our findings provide new insights into the functional role of CLU in breast cancer, highlighting a potential mechanistic basis by which CLU affects immune cell–tumor interactions. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1882/rc).
Methods
PrognoScan database (http://www.abren.net/PrognoScan/)
The PrognoScan database aims to facilitate a meta-analysis of the prognostic value of genes by comparing the relationship between gene expression and relevant outcomes, including overall survival (OS) in numerous published cancer microarray datasets (20). Accordingly, we used this database to evaluate the relationship between CLU expression and patient prognosis.
GEPIA database (http://gepia.cancer-pku.cn/index.html)
GEPIA is an online database based on web tools that provides customizable and rapid features based on Genotype-Tissue Expression (GTEX) and The Cancer Genome Atlas (TCGA) data. There are several features that make it interactive and customizable, including differential expression analysis, correlation analyses, mapping, similar genetic testing, patient survival analyses, and dimension reduction analysis (21). We utilized GEPIA database to evaluate the link between expression of CLU and patient prognosis and to further evaluate the link between expression of CLU and specific markers associated with tumor immune cell infiltration.
TIMER database (https://cistrome.shinyapps.io/timer/)
TIMER is a database for investigating immune cell infiltration in many cancers. In the database, a variety of statistical methods validated by pathological examination are used to analyze tumor infiltration by neutrophils, macrophages, DCs, B cells, and CD4/CD8 T cells (22). We first used this database to assess differences in CLU expression levels in specific tumor types and then explored the association between CLU expression and extent of infiltration by specific immune cell subsets. We performed Kaplan-Meier curve analysis to explore the impact of immune cell infiltration or gene expression on patient survival. In addition, we considered whether CLU expression correlated with specific markers of immune infiltrating cell subsets.
Kaplan-Meier plotter (http://kmplot.com/analysis/)
The Kaplan-Meier plotter provides a convenient method for exploring the impact of many different genes on survival of tumor patients with large sample sizes, including breast, ovarian, lung and gastric cancers (23). Based on this database, we investigated the association between CLU expression and prognosis in patients with these cancers, and the impact of clinicopathological factors and cancer-related outcomes. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
As part of the respective analyses, survival plots were generated based on the PrognoScan, TIMER, Kaplan-Meier plotter and GEPIA databases, with data such as hazard ratios (HRs) and P values, or P values derived from the log-rank test. Spearman’s correlation was used to measure the degree of correlation between specific variables, and the following R values were used to determine the degree of correlation. The range of 0.00–0.19 represented very weak, 0.20–0.39 weak, 0.40–0.59 moderate, 0.60–0.79 strong, and 1.0 extremely strong. We set a significance level of P<0.05.
Results
Assessment of CLU expression differences between tumors and normal tissues
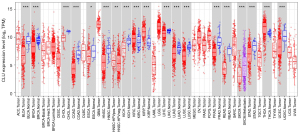
We evaluated differences in expression of CLU in multiple tumor types using the TIMER and TCGA databases. In comparison with normal control subjects, CLU was significantly higher in kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), and thyroid carcinoma (THCA). However, CLU expression was significantly reduced in these tumors compared to normal tissues, including bladder urothelial carcinoma (BLCA), cholangiocarcinoma (CHOL), breast invasive carcinoma (BRCA), esophageal carcinoma (ESCA), colon adenocarcinoma (COAD), kidney chromophobe (KICH), head and neck squamous cell carcinoma (HNSC), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), rectal adenocarcinoma (READ) and uterine corpus endometrial carcinoma (UCEC). Figure 1 illustrates a comparison of CLU expression in tumors and adjacent normal tissues from the TCGA dataset.
Relationship between CLU expression and prognosis of cancer patients
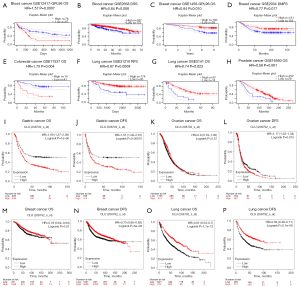
We used the PrognoScan database to study the relationship between CLU expression and prognosis of cancer patients (Figures S1-S4). We found that CLU expression was significantly correlated with prognosis of patients, including patients with hematological, breast, colon, lung and prostate cancers (Figure 2A-2H). We also used the Kaplan-Meier plotter database to evaluate the relationship between CLU expression and prognosis of these cancer types (Figure 2I-2P). The increase in CLU expression was significantly correlated with the poor prognosis of gastric cancer [OS HR =1.69, 95% confidence interval (CI): 1.37–2.09, P=1E-06; disease-free survival (DFS) HR =1.61, 95% CI: 1.26–2.07, P=1.5E-04]. However, the decrease in CLU expression was associated with poor prognosis in breast cancer [OS HR =0.78, 95% CI: 0.64–0.94, P=0.01; DFS HR =0.73, 95% CI: 0.65–0.82, P=5E-08] and lung cancer (OS HR =0.61, 95% CI: 0.53–0.7, P=1.1E-12; progression-free survival HR =0.59, 95% CI: 0.49–0.71, P=5.1E-08). There was no association between CLU expression and OS in ovarian cancer, but decreased expression improved DFS (OS HR =0.9, 95% CI: 0.78–1.03, P=0.12; DFS HR =1.17, 95% CI: 1.03–1.34, P=0.015). Using the GEPIA database, 33 TCGA tumor types were further analyzed to assess the correlation between CLU expression and patient outcomes and found that CLU expression correlated with OS in brain lower grade glioma (LGG), KIRC, pancreatic adenocarcinoma (PAAD), sarcoma (SARC), THCA and LIHC (figure available at https://cdn.amegroups.cn/static/public/tcr-22-1882-1.pdf). These results suggest that CLU expression is associated with different prognosis of multiple tumor types.
Correlation between CLU expression and prognosis of breast cancer patients with different molecular types
Since we found that CLU expression was associated with good prognosis in breast cancer patients, we examined the relationship between CLU expression and the molecular subtypes among breast cancer patients using Kaplan-Meier plots. CLU expression was significantly correlated with OS, DFS and with patient estrogen receptor (ER) status (positive array), HER2 status (positive array), subtype St Gallen (luminal B), subtype PAM50 (basal), TP53 status (mutated) and lymph node status, but not for grade and Pietenpol subtype (Table 1). We found no significant correlation between CLU expression and HER2 positive status (St Gallen) by molecular subtype, suggesting that there was no crosstalk between CLU expression and HER2 pathway.
Table 1
| Clinicopathological characteristics | Overall survival | Progression-free survival | |||||
|---|---|---|---|---|---|---|---|
| N | HR (95% CI) | P | N | HR (95% CI) | P | ||
| ER status—IHC | |||||||
| Positive | 754 | 0.77 (0.56–1.08) | 0.1286 | 2,633 | 0.75 (0.64–0.87) | 0.0001 | |
| Negative | 520 | 1.20 (0.85–1.69) | 0.2929 | 1,190 | 0.74 (0.60–0.91) | 0.0050 | |
| ER status—array | |||||||
| Positive | 1,309 | 0.78 (0.60–1.00) | 0.0465 | 3,768 | 0.81 (0.72–0.91) | 0.0005 | |
| Negative | 570 | 0.84 (0.61–1.15) | 0.2744 | 1,161 | 0.80 (0.66–0.97) | 0.0260 | |
| PR status—IHC | |||||||
| Positive | 156 | 2.97 (1.40–6.29) | 0.003 | 926 | 1.22 (0.91–1.64) | 0.1926 | |
| Negative | 291 | 0.78 (0.48–1.27) | 0.3194 | 925 | 0.65 (0.50–0.83) | 0.0007 | |
| HER2 status—array | |||||||
| Positive | 420 | 0.67 (0.47–0.96) | 0.0296 | 882 | 0.79 (0.63–0.98) | 0.0311 | |
| Negative | 1,459 | 0.81 (0.65–1.02) | 0.0759 | 4,047 | 0.77 (0.68–0.87) | 1.70E-05 | |
| Subtype St Gallen | |||||||
| Basal | 404 | 0.78 (0.53–1.15) | 0.2046 | 846 | 0.77 (0.61–0.97) | 0.0288 | |
| Luminal A | 794 | 1.58 (1.13–2.22) | 0.0072 | 2,277 | 0.91 (0.75–1.10) | 0.3202 | |
| Luminal B | 515 | 0.66 (0.46–0.95) | 0.0239 | 1,491 | 0.77 (0.64–0.92) | 0.0037 | |
| HER2+ | 166 | 1.38 (0.78–2.45) | 0.2706 | 315 | 0.79 (0.55–1.14) | 0.2074 | |
| Subtype PAM50 | |||||||
| Basal | 431 | 0.52 (0.35–0.77) | 0.0009 | 953 | 0.73 (0.58–0.91) | 0.0055 | |
| Luminal A | 596 | 1.56 (1.00–2.43) | 0.049 | 1,809 | 1.16 (0.92–1.47) | 0.2159 | |
| Luminal B | 439 | 1.16 (0.81–1.67) | 0.4068 | 1,353 | 0.83 (0.70–0.99) | 0.0358 | |
| HER2+ | 362 | 1.41 (0.96–2.07) | 0.0814 | 695 | 0.87 (0.68–1.12) | 0.2696 | |
| Normal | 51 | 2.08 (0.77–5.61) | 0.1384 | 119 | 0.72 (0.34–1.50) | 0.3777 | |
| Lymph node status | |||||||
| Positive | 452 | 0.70 (0.51–0.98) | 0.0348 | 1,656 | 0.63 (0.53–0.74) | 4.70E-08 | |
| Negative | 726 | 1.51 (1.02–2.25) | 0.0402 | 2,368 | 0.79 (0.67–0.92) | 0.0033 | |
| Grade | |||||||
| 1 | 175 | 1.67 (0.67–4.15) | 0.2658 | 397 | 1.42 (0.85–2.37) | 0.1753 | |
| 2 | 443 | 1.49 (0.98–2.28) | 0.0615 | 1,177 | 0.67 (0.53–0.84) | 0.0007 | |
| 3 | 586 | 1.25 (0.89–1.76) | 0.1902 | 1,300 | 0.87 (0.71–1.06) | 0.1545 | |
| TP53 status | |||||||
| Mutated | 130 | 0.51 (0.26–1.01) | 0.0486 | 188 | 0.52 (0.31–0.86) | 0.0091 | |
| Wild type | 197 | 0.73 (0.39–1.38) | 0.3298 | 273 | 0.78 (0.50–1.23) | 0.2861 | |
| Pietenpol subtype | |||||||
| Basal-like 1 | 103 | 0.24 (0.07–0.81) | 0.0126 | 251 | 0.69 (0.44–1.07) | 0.0982 | |
| Basal-like | 58 | 2.23 (0.77–6.44) | 0.1266 | 101 | 0.76 (0.40–1.42) | 0.3868 | |
| Immunomodulatory | 149 | 0.68 (0.31–1.50) | 0.3399 | 300 | 1.40 (0.88–2.22) | 0.1558 | |
| Mesenchymal | 114 | 0.72 (0.37–1.43) | 0.3517 | 211 | 0.78 (0.52–1.17) | 0.2296 | |
| Mesenchymal Stem-like | 39 | 1.76 (0.60–5.20) | 0.2989 | 81 | 1.48 (0.68–3.22) | 0.3229 | |
| Luminal androgen receptor | 116 | 1.22 (0.60–2.24) | 0.5164 | 253 | 0.84 (0.58–1.23) | 0.3743 | |
HR, hazard ratio; CI, confidence interval; IHC, immunohistochemistry; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TP53, tumor protein 53; PAM50, prediction analysis of microarray 50.
CLU expression correlates with the infiltration of breast cancer by immune cells
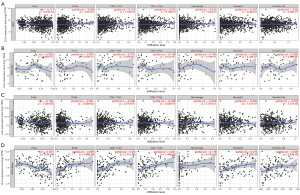
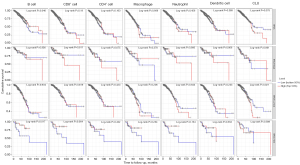
Prior studies have shown that the extent of immune cell infiltration impacts tumor prognosis in a variable way (24,25), especially for breast cancer (26,27). Therefore, we used the TIMER database to examine the relationship between CLU expression and immune cell infiltration across multiple tumor types (figure available at https://cdn.amegroups.cn/static/public/tcr-22-1882-2.pdf). CLU expression was significantly associated with tumor purity in 21 tumor types, and with B cell infiltration in 22 tumor types. CLU was also correlated with the level of CD8+ T cell infiltration in 10 tumor types, CD4+ T cell infiltration in 23 tumor types, macrophage infiltration in 21 tumor types, neutrophil infiltration in 12 tumor types, and DC infiltration in 20 tumor types. Within BRCA, BRCA luminal, and HER2 subtypes, CLU levels were not significantly associated with B cell, CD4+ T cell, CD8+ T cell, macrophage, neutrophil or DC infiltration (Figure 3A-3C). However, CLU expression was significantly correlated with the level of tumor purity (R=−0.301, P=5.3E-04), B cells (R=0.308, P=4.99E-04), CD8+ T cells (R=0.26, P=3.71E-03), CD4+ T cells (R=0.221, P=1.46E-02), macrophages (R=0.179, P=4.37E-02), neutrophils (R=0.275, P=3.70E-03) and DCs (R=0.205, P=2.84E-02) in BRCA basal type (Figure 3D). In BRCA basal type, CLU expression was significantly correlated with immune cell infiltration, especially B cells. This is consistent with previous results (28,29). We further used the TIMER database to generate Kaplan-Meier plots to investigate the correlation between CLU expression and immune cell infiltration in BRCA and its subtypes. B-cell infiltration and CLU expression were significantly associated with prognosis in BRCA (P=0.046) and HER2 (P=0.017) subtypes but not in BRCA basal and luminal subtypes (Figure 4). This suggests that CLU regulates the infiltration of immune cells in breast cancer.
Correlation between CLU expression and immune markers
Using the TIMER and GEPIA databases, we investigated further the connection between CLU expression and immune cell infiltration based on immune marker sets in BRCA. We examined the association between CLU expression and markers of specific cell subsets, including total T cells, CD8+ T cells, M1/M2 macrophages, B cells, natural killer (NK) cells, monocytes, neutrophils, DCs, TAMs, Th1 cells, Th2 cells, Th17 cells, T follicular helper (Tfh) cells, Treg cells and exhausted T cells. After adjusting for tumor purity, CLU expression correlated with TAM markers (CD68 and IL-10), monocyte markers (CD115), DC markers (HLA-DPB1 and HLA-DQB1), M1 macrophage markers [inducible nitric oxide synthase (iNOS) and IRF5], NK cell markers (KIR3DL3), Th1 markers [interferon γ (IFN-γ)], Th2 markers (GATA3, STAT6 and STAT5A), Th17 markers in BRCA (STAT3), Tfh markers (BCL6), Treg cell markers (Foxp3, CCR8 and STAT5b) and T cell exhaustion markers [cytotoxic T-lymphocyte-associated protein 4 (CTLA4), lymphocyte activation gene 3 (LAG3) and GZMB] (Table 2). There was a significant correlation between CLU expression and DC markers (HLA-DPB1 and HLA-DQB1), Th2 markers (GATA3, STAT6 and STAT5A), Treg cell markers (FOXP3, CCR8 and STAT5B) and T cell exhaustion markers (CTLA4 and LAG3) in BRCA (P<0.05; Figure 5). Therefore, we further evaluated the relationship between CLU expression and these markers in BRCA using the GEPIA database, which revealed similar correlations between CLU and markers of DCs, Th2 cells, Treg cells and T cell exhaustion, and between CLU expression in TIMER and these markers (Table 3). This suggested that elevated CLU expression in BRCA increased DC infiltration, and expression of DC markers HLA-DQB1, HLA-DPB1, HLA-DPA1, HLA-DRA, BDCA-4 and BDCA-1 correlated with CLU expression. CLU was closely related to tumor DC penetration. DCs can promote tumor metastasis by enhancing Treg cell responses and suppressing CD8+ T cell cytotoxicity (30). Further work is required to determine whether CLU plays a critical role in regulating DC infiltration and tumor metastasis. We further observed a significant correlation between CLU, T-cell subsets, including Foxp3 CCR8, STAT5b, transforming growth factor β (TGFβ), CTLA4 and LAG3 (Table 2), suggesting that CLU may play an immune escape role in the progression of breast cancer, although the mechanism needs to be confirmed.
Table 2
| Description | Gene makers | BRCA | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| Cor | P | Cor | P | |||
| CD8+ T cell | CD8A | 0.025 | 0.416 | −0.037 | 0.242 | |
| CD8B | 0 | 0.997 | −0.054 | 0.090 | ||
| T cell (general) | CD3D | 0.013 | 0.655 | −0.054 | 0.089 | |
| CD3E | 0.022 | 0.473 | −0.047 | 0.137 | ||
| CD2 | 0 | 0.998 | −0.068 | 0.033 | ||
| B cell | CD19 | 0.024 | 0.421 | −0.033 | 0.297 | |
| CD79A | 0.03 | 0.327 | −0.034 | 0.289 | ||
| Monocyte | CD86 | −0.009 | 0.765 | −0.056 | 0.080 | |
| CD115 (CSF1R) | 0.103 | 5.87E-04 | 0.063 | 0.047 | ||
| TAM | CCL2 | 0.012 | 0.68 | −0.033 | 0.298 | |
| CD68 | −0.026 | 0.387 | −0.076 | 0.017 | ||
| IL10 | −0.035 | 0.247 | −0.081 | 0.010 | ||
| M1 macrophage | INOS (NOS2) | −0.072 | 0.0163 | −0.078 | 0.014 | |
| IRF5 | 0.127 | 2.38E-05 | 0.105 | 9.49E-04 | ||
| COX2 (PTGS2) | 0.038 | 0.0202 | −0.003 | 0.932 | ||
| M2 macrophage | CD163 | −0.005 | 0.872 | −0.047 | 0.141 | |
| VSIG4 | 0.052 | 0.0864 | 0.011 | 0.741 | ||
| MS4A4A | 0.021 | 0.494 | −0.03 | 0.340 | ||
| Neutrophils | CD66b (CEACAM8) | 0.016 | 0.586 | 0.01 | 0.742 | |
| CD11b (ITGAM) | 0.047 | 0.118 | 0.018 | 0.562 | ||
| CCR7 | 0.033 | 0.286 | −0.027 | 0.391 | ||
| NK cells | KIR2DL1 | −0.003 | 0.927 | −0.031 | 0.322 | |
| KIR2DL3 | 0.011 | 0.718 | −0.018 | 0.561 | ||
| KIR2DL4 | 0.009 | 0.761 | −0.022 | 0.482 | ||
| KIR3DL1 | 0.011 | 0.717 | −0.02 | 0.525 | ||
| KIR3DL2 | 0.029 | 0.345 | −0.024 | 0.459 | ||
| KIR3DL3 | −0.035 | 0.24 | −0.073 | 0.022 | ||
| KIR2DS4 | 0.008 | 0.797 | −0.021 | 0.499 | ||
| DC | HLA-DPB1 | 0.131 | 1.22E-05 | 0.091 | 3.95E-03 | |
| HLA-DQB1 | 0.092 | 2.19E-03 | 0.065 | 4.02E-02 | ||
| HLA-DRA | 0.06 | 4.54E-02 | 0.011 | 0.740 | ||
| HLA-DPA1 | 0.102 | 6.96E-04 | 0.061 | 5.48E-02 | ||
| BCDA-1 (CD1C) | 0.096 | 1.41E-03 | 0.039 | 0.221 | ||
| BDCA-4 (NRP1) | 0.079 | 8.52E-03 | 0.031 | 0.334 | ||
| CD11c (ITGAX) | 0.009 | 0.759 | −0.039 | 0.217 | ||
| Th1 cell | T-bet (TBX21) | 0.022 | 0.476 | −0.042 | 0.181 | |
| STAT4 | 0.045 | 0.134 | −0.009 | 0.774 | ||
| STAT1 | −0.035 | 0.246 | −0.054 | 9.07E-02 | ||
| IFN-γ (IFNG) | −0.038 | 0.21 | −0.091 | 3.97E-03 | ||
| TNF-α (TNF) | −0.026 | 0.38 | −0.029 | 0.368 | ||
| Th2 cell | GATA3 | 0.126 | 2.87E-05 | 0.153 | 1.21E-06 | |
| STAT6 | 0.194 | 9.09E-11 | 0.181 | 8.58E-09 | ||
| STAT5A | 0.217 | 3.33E-13 | 0.202 | 1.38E-10 | ||
| IL13 | −0.028 | 0.355 | −0.042 | 0.186 | ||
| Tfh cell | BCL6 | 0.151 | 5.14E-07 | 0.147 | 3.11E-06 | |
| IL21 | −0.025 | 0.41 | −0.049 | 0.119 | ||
| Th17 cell | STAT3 | 0.12 | 6.81E-05 | 0.117 | 2.10E-04 | |
| IL17A | 0.048 | 0.109 | 0.019 | 0.557 | ||
| Treg cell | FOXP3 | −0.063 | 3.62E-02 | −0.118 | 2.04E-04 | |
| CCR8 | −0.078 | 1.01E-02 | −0.118 | 2.03E-04 | ||
| STAT5B | 0.185 | 6.38E-10 | 0.168 | 1.04E-07 | ||
| TGFβ (TGFB1) | 0.084 | 5.36E-03 | 0.041 | 0.202 | ||
| T cell exhaustion | PD-1 (PDCD1) | 0.023 | 0.447 | −0.032 | 0.307 | |
| CTLA4 | −0.063 | 3.68E-02 | −0.12 | 1.42E-04 | ||
| LAG3 | −0.061 | 4.18E-02 | −0.096 | 2.37E-03 | ||
| TIM-3 (HAVCR2) | 0.003 | 0.92 | −0.038 | 0.230 | ||
| GZMB | −0.032 | 0.287 | −0.087 | 5.91E-03 | ||
Cor, R value of Spearman’s correlation; None, correlation without adjustment; Purity, correlation adjusted by purity; BRCA, breast invasive carcinoma; CD, cluster of differentiation; TAM, tumor associated macrophage; CCL2, chemokine ligand 2; IL, interleukin; INOS, inducible nitric oxide synthase; IRF, interferon regulatory factor; COX2, cyclooxygenase 2; VSIG4, V-set and immunoglobulin domain-containing protein 4; CCR, chemokine receptor; KIR2DL1, killer cell immunoglobulin-like receptor 2DL1; NK cell, natural killer cell; DC, dendritic cell; HLA, human leukocyte antigen; STAT4, signal transducer and activator of transcription 4; IFN, interferon; FOXP3, forkhead box p3; TGF-β, transforming growth factor beta; CTLA4, cytotoxic T-lymphocyte-associated protein 4; LAG3, lymphocyte activation gene-3; TIM-3, T cell immunoglobulin and mucin domain-containing protein 3.
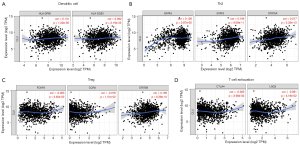
Table 3
| Description | Gene makers | BRCA | ||||
|---|---|---|---|---|---|---|
| Tumor | Normal | |||||
| R | P | R | P | |||
| DCs | HLA-DPB1 | 0.130 | 1.20E-05 | 0.460 | 4.10E-07 | |
| HLA-DQB1 | 0.060 | 0.049 | 0.290 | 2.20E-03 | ||
| HLA-DRA | 0.064 | 0.036 | 0.270 | 3.80E-03 | ||
| HLA-DPA1 | 0.110 | 3.70E-04 | 0.220 | 0.021 | ||
| BCDA-1 (CD1C) | 0.086 | 4.50E-03 | 0.220 | 0.019 | ||
| BDCA-4 (NRP1) | 0.130 | 1.40E-05 | 0.120 | 0.200 | ||
| CD11c (ITGAX) | 0.025 | 0.410 | 0.310 | 0.001 | ||
| Th2 cells | GATA3 | 0.150 | 3.20E-07 | 0.062 | 0.520 | |
| STAT6 | 0.210 | 3.70E-12 | 0.170 | 0.070 | ||
| STAT5A | 0.230 | 5.20E-15 | −0.074 | 0.440 | ||
| IL13 | 0.018 | 0.560 | 9.10E-03 | 0.920 | ||
| Treg cells | FOXP3 | −0.069 | 0.024 | 0.220 | 0.020 | |
| CCR8 | −0.047 | 0.120 | 0.120 | 0.220 | ||
| STAT5B | 0.220 | 2.00E-13 | −0.058 | 0.540 | ||
| TGFβ (TGFB1) | 0.095 | 1.70E-03 | 0.460 | 3.30E-07 | ||
| T cell exhaustion | PD-1 (PDCD1) | 0.011 | 0.710 | 0.290 | 1.80E-03 | |
| CTLA4 | −0.059 | 0.054 | −0.065 | 0.490 | ||
| LAG3 | −0.082 | 7.10E-03 | 0.150 | 0.130 | ||
| TIM-3 (HAVCR2) | 0.028 | 0.360 | 0.350 | 1.40E-04 | ||
| GZMB | −0.048 | 0.120 | 0.340 | 2.30E-04 | ||
CLU, clusterin; GEPIA, Gene Expression Profiling Interactive Analysis; BRCA, breast cancer; IL, interleukin; CCR, chemokine receptor; DCs, dendritic cells; HLA, human leukocyte antigen; CD, cluster of differentiation; STAT, signal transducer and activator of transcription; FOXP3, forkhead box p3; CTLA4, cytotoxic T-lymphocyte-associated protein 4; LAG3, lymphocyte activation gene-3; TIM-3, T cell immunoglobulin and mucin domain-containing protein 3.
Discussion
CLU is a secretory glycoprotein and is essentially a heterodimer. It is expressed in a variety of tissues and body fluids. CLU is also considered to be a promising biomarker for cell death, malignancy, cancer progression and drug resistance development (31). CLU plays an important carcinogenic role by promoting various downstream carcinogenic pathways (11,32-34). Protein kinase D3 is a key regulator of CLU and promoted tumor growth in triple-negative breast cancer (35). In HER2-positive breast cancer, trastuzumab treatment upregulates expression of CLU protein, which is positively correlated with the dose. By blocking the CLU expression induced by trastuzumab, OGX-011 treatment might enhance the growth inhibitory effect of monoclonal antibody trastuzumab (36). In this study, we found that in several types of cancer, CLU expression was correlated with the prognosis of patients, and low CLU expression was strongly correlated with poor prognosis of BRCA. BRCA patients with low CLU expression are also more likely to be ER and HER2 negative, suggesting that CLU may be useful as a prognostic indicator. We found that expression of CLU in tumors was associated with many different markers of immune cell subsets, which highlighted the possible role of CLU in the immune interaction between BRCA and such tumors, making CLU a promising biomarker for further investigation. We evaluated the expression of CLU using an independent GEPIA database because it is related to the prognosis of many different types of cancer. In these cancers, the expression of CLU in tumor tissues was significantly different from that in normal tissues. TCGA data set analysis indicated that there was elevated CLU expression in KIRC, KIRP and THCA, whereas expression was decreased in BLCA, BRCA, CHOL, COAD, ESCA, HNSC, LUAD, KICH, LIHC, LUSC, READ, PRAD, STAD and UCEC relative to adjacent control tissues. In a series of different cancers, depending on what method was used in the study, or what mechanism was involved, CLU expression may have changed. In these databases, we consistently observed that decreased CLU expression was associated with poor prognosis of BRCA. In the TCGA database, elevated CLU levels were associated with poor prognosis in LGG patients, while LIHC results were the opposite. Similarly, the Kaplan-Meier database found that the decrease in CLU was associated with poor prognosis of breast and lung cancers. Decreased expression of CLU was associated with poorer prognosis, as well as ER status (array), HER2 status (array), subtype, and lymph node status. These results suggest that CLU may be a valuable biomarker for the prognosis of BRCA.
The expression of CLU is also correlated with immune infiltration in many cancers, including BRCA. We found that expression of CLU was weakly positively correlated with infiltration of B cells, CD8+ T cells, CD4+ T cells, DCs and neutrophils in BRCA. We further found that B cell infiltration was significantly correlated with the prognosis of BRCA. The correlation between CLU and expression of some immune marker genes strongly suggests that CLU can control the infiltration and interaction of immune cells in the tumor microenvironment in BRCA. We observed a weak correlation between CLU and Th2 marker STAT5A. This suggests that CLU regulates humoral immunity. We further found that CLU levels in BRCA were associated with markers of Treg cells and T cell failure (CTLA4 and LAG3). It is suggested that CLU can promote Treg cell response and inhibit T-cell immunity. We found that expression of CLU is linked to expression of multiple T cell markers (Th2, Tfh and Th17) in BRCA. This may reflect that CLU is involved in regulating T cell response in BRCA, and suggests that CLU plays a role in regulating the recruitment and activation of immune cells in BRCA.
In conclusion, CLU may play an important regulatory role in tumor immune cell infiltration, and is also a valuable prognostic biomarker for patients with breast cancer.
Acknowledgments
We would like to thank all the researchers who participated in this study.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1882/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1882/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1882/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Puvirajesinghe TM, Bertucci F, Jain A, et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nat Commun 2016;7:10318. [Crossref] [PubMed]
- Chen N, Higashiyama N, Hoyos V. Predictive Biomarkers of Immune Checkpoint Inhibitor Response in Breast Cancer: Looking beyond Tumoral PD-L1. Biomedicines 2021; [Crossref] [PubMed]
- Miles D, Gligorov J, André F, et al. Primary results from IMpassion131, a double-blind, placebo-controlled, randomised phase III trial of first-line paclitaxel with or without atezolizumab for unresectable locally advanced/metastatic triple-negative breast cancer. Ann Oncol 2021;32:994-1004. [Crossref] [PubMed]
- Emens LA, Adams S, Barrios CH, et al. First-line atezolizumab plus nab-paclitaxel for unresectable, locally advanced, or metastatic triple-negative breast cancer: IMpassion130 final overall survival analysis. Ann Oncol 2021;32:983-93. [Crossref] [PubMed]
- Cortes J, Cescon DW, Rugo HS, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020;396:1817-28. [Crossref] [PubMed]
- Skala MC, Ayuso JM, Burkard ME, et al. Breast cancer immunotherapy: current biomarkers and the potential of in vitro assays. Curr Opin Biomed Eng 2021;21:100348. [Crossref] [PubMed]
- Waniczek D, Lorenc Z, Śnietura M, et al. Tumor-Associated Macrophages and Regulatory T Cells Infiltration and the Clinical Outcome in Colorectal Cancer. Arch Immunol Ther Exp (Warsz) 2017;65:445-54. [Crossref] [PubMed]
- Praharaj PP, Patra S, Panigrahi DP, et al. Clusterin as modulator of carcinogenesis: A potential avenue for targeted cancer therapy. Biochim Biophys Acta Rev Cancer 2021;1875:188500. [Crossref] [PubMed]
- Peng M, Deng J, Zhou S, et al. The role of Clusterin in cancer metastasis. Cancer Manag Res 2019;11:2405-14. [Crossref] [PubMed]
- Tan J, Guo W, Yang S, et al. The multiple roles and therapeutic potential of clusterin in non-small-cell lung cancer: a narrative review. Transl Lung Cancer Res 2021;10:2683-97. [Crossref] [PubMed]
- Patarat R, Riku S, Kunadirek P, et al. The expression of FLNA and CLU in PBMCs as a novel screening marker for hepatocellular carcinoma. Sci Rep 2021;11:14838. [Crossref] [PubMed]
- Fu N, Du H, Li D, et al. Clusterin contributes to hepatitis C virus-related hepatocellular carcinoma by regulating autophagy. Life Sci 2020;256:117911. [Crossref] [PubMed]
- Ma J, Gao W, Gao J. sCLU as prognostic biomarker and therapeutic target in osteosarcoma. Bioengineered 2019;10:229-39. [Crossref] [PubMed]
- Rodríguez-Rivera C, Garcia MM, Molina-Álvarez M, et al. Clusterin: Always protecting. Synthesis, function and potential issues. Biomed Pharmacother 2021;134:111174. [Crossref] [PubMed]
- Afanasyeva MA, Britanova LV, Korneev KV, et al. Clusterin is a potential lymphotoxin beta receptor target that is upregulated and accumulates in germinal centers of mouse spleen during immune response. PLoS One 2014;9:e98349. [Crossref] [PubMed]
- Merlotti A, Dantas E, Remes Lenicov F, et al. Fucosylated clusterin in semen promotes the uptake of stress-damaged proteins by dendritic cells via DC-SIGN. Hum Reprod 2015;30:1545-56. [Crossref] [PubMed]
- Niu ZH, Wang Y, Chun B, et al. Secretory clusterin (sCLU) overexpression is associated with resistance to preoperative neoadjuvant chemotherapy in primary breast cancer. Eur Rev Med Pharmacol Sci 2013;17:1337-44. [PubMed]
- Wang Y, Wang X, Zhao H, et al. Clusterin confers resistance to TNF-alpha-induced apoptosis in breast cancer cells through NF-kappaB activation and Bcl-2 overexpression. J Chemother 2012;24:348-57. [Crossref] [PubMed]
- Mizuno H, Kitada K, Nakai K, et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2009;2:18. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Lánczky A, Nagy Á, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 2016;160:439-46. [Crossref] [PubMed]
- Hennequin A, Derangère V, Boidot R, et al. Tumor infiltration by Tbet+ effector T cells and CD20+ B cells is associated with survival in gastric cancer patients. Oncoimmunology 2015;5:e1054598. [Crossref] [PubMed]
- Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the 'Immunoscore' in the classification of malignant tumours. J Pathol 2014;232:199-209. [Crossref] [PubMed]
- Lee KH, Kim EY, Yun JS, et al. The prognostic and predictive value of tumor-infiltrating lymphocytes and hematologic parameters in patients with breast cancer. BMC Cancer 2018;18:938. [Crossref] [PubMed]
- Fan Z, Yu P, Wang Y, et al. NK-cell activation by LIGHT triggers tumor-specific CD8+ T-cell immunity to reject established tumors. Blood 2006;107:1342-51. [Crossref] [PubMed]
- Cao Y, Chen C, Tao Y, et al. Immunotherapy for Triple-Negative Breast Cancer. Pharmaceutics 2021; [Crossref] [PubMed]
- Fan Y, He S. The Characteristics of Tumor Microenvironment in Triple Negative Breast Cancer. Cancer Manag Res 2022;14:1-17. [Crossref] [PubMed]
- Nomura T, Yamakawa M, Shimaoka T, et al. Development of Dendritic Cell-Based Immunotherapy Targeting Tumor Blood Vessels in a Mouse Model of Lung Metastasis. Biol Pharm Bull 2019;42:645-8. [Crossref] [PubMed]
- Das L, Shekhar S, Chandrani P, et al. In silico structural analysis of secretory clusterin to assess pathogenicity of mutations identified in the evolutionarily conserved regions. J Biomol Struct Dyn 2021; Epub ahead of print. [Crossref] [PubMed]
- Tian Y, Wang C, Chen S, et al. Extracellular Hsp90α and clusterin synergistically promote breast cancer epithelial-to-mesenchymal transition and metastasis via LRP1. J Cell Sci 2019;132:jcs228213. [Crossref] [PubMed]
- Mitsufuji S, Iwagami Y, Kobayashi S, et al. Inhibition of Clusterin Represses Proliferation by Inducing Cellular Senescence in Pancreatic Cancer. Ann Surg Oncol 2022;29:4937-46. [Crossref] [PubMed]
- Zhao W, Wang X, Jiang Y, et al. miR-217-5p Inhibits Invasion and Metastasis of Prostate Cancer by Targeting Clusterin. Mamm Genome 2021;32:371-80. [Crossref] [PubMed]
- Liu Y, Zhou Y, Ma X, et al. Inhibition Lysosomal Degradation of Clusterin by Protein Kinase D3 Promotes Triple-Negative Breast Cancer Tumor Growth. Adv Sci (Weinh) 2021;8:2003205. [Crossref] [PubMed]
- Biroccio A, D'Angelo C, Jansen B, et al. Antisense clusterin oligodeoxynucleotides increase the response of HER-2 gene amplified breast cancer cells to Trastuzumab. J Cell Physiol 2005;204:463-9. [Crossref] [PubMed]


