
Knockdown of MEF2D inhibits the development and progression of B-cell acute lymphoblastic leukemia
Highlight box
Key findings
• MEF2D is overexpressed in B-ALL patients.
• MEF2D promotes tumor progression of B-ALL in vitro and in vivo, which may be related to the PI3K-AKT signaling pathway.
What is known and what is new?
• MEF2D is involved in the progression of various malignant tumors.
• MEF2D plays a vital role in the development and progression of B-ALL.
What are the implications, and what should change now?
• MEF2D may be a potential novel target for B-ALL therapy.
Introduction
Acute lymphoblastic leukemia (ALL) is a hematologic malignancy characterized by the malignant multiplication of lymphoid progenitor cells in the bone marrow, peripheral blood, and extramedullary sites. According to clinical statistics, leukemia accounts for 4% and 3% of oncological diseases in males and females, respectively, with mortality rates of 4% and 3%, respectively (1). The incidence of B-cell ALL (B-ALL) accounts for 85% and 75% of ALL cases in children and adults, respectively (2). Recently, frontline regimens for ALL treatment have resulted in a 5-year overall survival (OS) rate of 90% in pediatric patients (3,4) but less than 45% in adults (4-6). In older patients (>60 years), the 5-year OS rate is under 20% (7). In addition, approximately one-third of patients with standard-risk ALL and two-thirds of patients with high-risk ALL will experience relapse (8). Hematopoietic stem cell transplantation remains the most potent therapeutic approach for B-ALL; however, some patients are not eligible for transplantation. Furthermore, transplantation-related complications and post-transplantation relapse are still clinical obstacles. Therefore, there is an urgent need to explore novel therapeutic targets and new therapeutic strategies to combat B-ALL.
Myocyte enhancer factor 2D (MEF2D) is a member of the MEF2 protein family, which is a transcription factor (TF) family containing MEF2A, MEF2B, MEF2C, and MEF2D, and plays cellular functions in skeletal, cardiac, neuronal, and hematopoietic development (9). Notably, it has been extensively demonstrated that MEF2D is overexpressed in a diversity of neoplasms, such as hepatocellular carcinoma, pancreatic cancer, and breast cancer, and is significantly correlated with tumor progression and adverse prognosis (10-12). A retroviral insertional analysis performed in a murine model has identified MEF2D as a candidate gene involved in leukemogenesis (13). A previous study showed that MEF2D is overexpressed in leukemia patients (14). Furthermore, chromosomal translocations involving MEF2D have been reported in B-ALL cases, and rearrangements result in enhanced MEF2D transcriptional activity and the arrest of B-cell differentiation, thus contributing to the development of a distinct subtype of high-risk leukemia (15). This makes MEF2D a potential therapeutic target for B-ALL. However, the function and molecular mechanism of MEF2D in B-ALL remain unclear.
In this study, we revealed that MEF2D was aberrantly overexpressed in B-ALL and that MEF2D overexpression was remarkably correlated to disease progression in ALL patients. Also, MEF2D knockdown in B-ALL cells significantly restrained cell viability, induced cell apoptosis, arrested cell cycle progression, and enhanced drug resistance in vitro. Using a B-ALL xenograft mouse model, we confirmed that MEF2D knockdown markedly reduced the B-ALL tumor load in vivo. Additionally, we also elucidated that the knockdown of MEF2D downregulated the expression of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)-protein kinase B (AKT) signaling pathway in B-ALL. Thus, MEF2D served as a potential therapeutic target in B-ALL. We present the following article in accordance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1778/rc).
Methods
Cell culture
Human B-ALL cell lines Nalm6 (RRID: CVCL_0092) and RS4;11 (RRID: CVCL_0093) were ordered from American Type Culture Collection (ATCC; Guang Zhou Jennio Biotech Co., Ltd., Guangzhou, China). The cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Biological Industries, Beit Haemek, Israel) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Israel) and 1% streptomycin and penicillin at 37 ℃ with 5% CO2.
Clinical samples
Bone marrow samples from B-ALL patients and healthy donors were acquired from Zhujiang Hospital of Southern Medical University (Guangzhou, China). This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the Zhujiang Hospital of Southern Medical University (No. 2022-KY-129). Informed consent was provided by all individual participants.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the fresh-frozen cells in Trizol Reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription of 1,000 ng RNA was conducted using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Shiga, Japan) in accordance with the manufacturer’s instructions. qRT-PCR in a final volume of 20 µL was conducted using TB Green® Premix Ex TaqTM II (Tli RNaseH Plus) (Takara) and the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). In each 20 µL reaction system, 1 µL of complementary DNA (cDNA) was mixed with the forward and reverse primers, TB Green® Premix Ex TaqTM II (Tli RNaseH Plus) and double distilled water (ddH2O). The relative expression of messenger RNA (mRNA) was evaluated by the 2−ΔΔCt method, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was considered as a normalization control. The PCR primer sequences are listed in Table 1. The amplification efficiency and correlation coefficient of PCR primers are listed in Table S1.
Table 1
| Gene | Sense primer | Antisense primer |
|---|---|---|
| MEF2D | AGGGAAATAACCAAAAAACTACCAAA | GCTACATGAACACAAAAACAGAGACC |
| PIK3CB | GCTGAACAGTAGCAATGTGGC | ATTCCTCAATGGCTCGGTCC |
| AKT | GACGGGCACATTAAGATCAC | TGAGGATGAGCTCAAAAAGC |
| BCL2 | CTTTGAGTTCGGTGGGGTCA | GGGCCGTACAGTTCCACAAA |
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
qRT-PCR, quantitative reverse transcription polymerase chain reaction; MEF2D, myocyte enhancer factor 2D; PIK3CB, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta; AKT, protein kinase B; BCL2, B-cell lymphoma-2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western blotting assay
According to the literature (16), cells were harvested and lysed by radioimmunoprecipitation assay (RIPA) lysis buffer containing 1% phosphatase inhibitor and 1% phenylmethanesulfonyl fluoride at 4 ℃ for 10 minutes. Then, the supernatants were harvested following centrifugation. Protein concentrations were quantified using the bicinchoninic acid (BCA) Protein Assay Kit (Beyotime, Beijing, China). After adding the loading buffer, the proteins were separated by electrophoresis in 10% sodium dodecyl sulfate (SDS) polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were then blocked in 5% non-fat milk/tris-buffered saline with Tween 20 (TBST) and incubated with respective primary antibodies overnight at 4 ℃. After washing in TBST 3 times, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary anti-mouse or anti-rabbit antibodies for 1 hour at room temperature. The protein bands were detected using an enhanced chemiluminescence (ECL) kit (FDbio Science, Hangzhou, China). The antibodies used were as follows: MEF2D (Rabbit, Polyclonal, 1:1,000, 14353-1-AP), GAPDH (Mouse, Monoclonal, 1:10,000, 60004-1-Ig), AKT (Rabbit, Polyclonal, 1:2,000, 10176-2-AP), phosphorylated (p)-AKT (Mouse, Monoclonal, 1:2,000, 66444-1-IG-50UL), B-cell lymphoma 2 (BCL2) (Rabbit, Polyclonal, 1:2,000, 12789-1-AP), HRP-conjugated secondary anti-mouse antibody (Goat, 1:10,000, SA00001-1), HRP-conjugated secondary anti-rabbit antibody (Goat, 1:10,000, SA00001-2) (both from Proteintech, Wuhan, China), and PI3K (Rabbit, Monoclonal, 1:1,000, 3011T, Cell Signaling Technology, Danvers, MA, USA).
Lentivirus production and B-ALL cells transduction
The lentivirus vector knockdown MEF2D gene [MEF2D-short hairpin (sh)RNA] and negative control (NC) vector were purchased from GeneChem Co., Ltd. (Shanghai, China). The vector, which contained a green fluorescent protein (GFP) and a puromycin-resistant gene, was packaged in lentivirus. Nalm6 and RS4;11 cells were individually transduced with MEF2D-shRNA and NC lentivirus at 50 multiplicity of infection for 72 hours. Next, puromycin was applied to perform the positive selection. After amplification, the efficiency of MEF2D knockdown was verified by qRT-PCR analysis and western blotting. Successfully transduced cells were available for subsequent experiments.
Cell viability assays
B-ALL cells were washed, seeded in 96-well plates, and cultured at a concentration of 1×104 cells per well. After incubation for 0, 24, 48, and 72 hours, a 10 µL Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Kumamoto, Japan) was added to each well. After incubating at 37 ℃ for 4 hours, a microplate reader (Multiskan Go; Thermo Fisher Scientific, USA) was used to validate the optical density (OD) of each well at 450 nM, and cell viability curves were plotted according to the OD values.
Cell apoptosis assays
Cells at a density of 1×105 were seeded into plates and harvested after 24 hours. To determine the degree of cell apoptosis, cells were incubated with the Annexin-V allophycocyanin (APC) & propidium iodide (PI) Apoptosis Detection Kit (Keygen Biotech, Nanjing, China) for 15 minutes at room temperature in the dark. Data were detected using the CytoFLEX flow cytometer (Beckman Coulter, Brea, CA, USA).
Cell cycle assays
The 1×106 cells were collected and washed twice with cold phosphate-buffered saline (PBS; Gibco, Waltham, MA, USA). Then, the cells were resuspended in the cold 70% ethanol and incubated overnight at 4 ℃. The following day, the fixed cells were washed twice with cold PBS and stained with 500 µL PI (BD Pharmingen, San Diego, CA, USA) for 15 minutes at room temperature in the dark. Data were detected using the CytoFLEX flow cytometer (Beckman Coulter, USA).
Drug sensitivity assays
The MEF2D-shRNA and NC cells (1×104 cells/well) were plated onto 96-well culture plates in the presence of different concentrations of either dexamethasone (DXMS; Sigma Aldrich, St. Louis, MO, USA), vincristine (VCR; Keygen Biotech, China), or venetoclax (MedChemExpress, Princeton, NJ, USA) for the treatment groups, and in the presence of a corresponding concentration of dimethyl sulfoxide (DMSO; MP Biomedicals, Santa Ana, CA, USA) for the vehicle control groups. At the indicated time points, the 10 µL CCK-8 assay was added to each well. After incubating at 37 ℃ for 4 hours, the cell viability percentage was evaluated by assessing the absorbance at 450 nm and normalized to the corresponding vehicle control.
The MEF2D-shRNA and NC cells (1×105 cells/well) were plated onto 24-well culture plates in the presence of either DXMS, VCR, or venetoclax for the treatment groups, and in the presence of a corresponding concentration of DMSO for the vehicle control groups. At the indicated time points, the cells were incubated with the Annexin-V APC & PI Apoptosis Detection Kit for 15 minutes at room temperature in the dark, followed by flow cytometry (FCM) analysis.
Xenograft tumor experiments
Balb/c nude mice (females, 4 weeks old, weight range: 14–16 g) were sourced from the Zhujiang Hospital Animal Center and housed in a specific pathogen-free (SPF) environment. The animal experiments were performed under a project license (No. LAEC-2020-117) granted by the Animal Ethics Committee of Zhujiang Hospital of Southern Medical University, in compliance with Zhujiang Hospital’s guidelines for the care and use of animals.
To establish the B-ALL xenograft animal model, NC of Nalm6 (Nalm6/NC) and MEF2D-knockdown Nalm6 (Nalm6/sh) cells were washed twice with PBS and adjusted to the density of 5×107 cells/mL and then injected subcutaneously (5×106 cells per mouse in 100 µL of PBS). Tumor growth was measured using a vernier caliper and body weight was measured by an electronic scale every 2 days. All mice were euthanized and sacrificed on the 26th day after injection. The volume of tumors was determined using the formula: (length × width2)/2. Tumor tissues were harvested, processed, and used in subsequent experiments. If the volume of the tumor decreased after inoculation and no longer increased, the sample was excluded.
Hematoxylin and eosin (H&E) staining and immunohistochemical (IHC) staining
Fresh tissues were fixed, embedded in paraffin wax, and subjected to H&E staining or IHC staining according to the established protocols. The sections were dewaxed in xylene and rehydrated in ethanol, and endogenous peroxidase was blocked by methanol containing 0.3% hydrogen peroxide for 30 minutes. For antigen retrieval, the sections were placed in plastic jars containing a citric acid antigen retrieval buffer (Servicebio, Wuhan, China) and then heated in a microwave oven. Non-specific protein binding was inhibited by treatment with normal goat serum (for rabbit polyclonal antibodies) (Servicebio) or 3% bovine serum albumin (BSA; Servicebio) for 30 minutes. Sections were incubated at 4 ℃ overnight with primary antibodies, and the tissues were covered with a secondary antibody (HRP-labeled; Servicebio) from the corresponding species of primary antibody and incubated at room temperature. The sections were then reacted with a 3,3'-diaminobenzidine (DAB) color-developing solution (Servicebio, China), counterstained with hematoxylin, and mounted. The primary antibodies used were as follows: MEF2D (Proteintech, China), PI3K (Proteintech, China), p-AKT (Proteintech, China), and BCL2 (Proteintech, China).
RNA sequencing analysis
A quantity of 200 ng of total RNA was isolated and used for RNA sequencing analysis. The RNA sequencing analysis was then carried out via a commercially available service (BGI-Huada Gene, Shenzhen, China). Briefly, after total RNA was fragmented into short fragments and mRNA was enriched using Library Preparation VAHTS mRNA Capture Beads (Invitrogen), double-stranded cDNA was purified and enriched by PCR amplification. Next, a cDNA library was constructed using an MGIEasy RNA Library Prep Kit (MGI, Shenzhen, China) and cDNA products were sequenced using BGIseq-500 (BGI-Huada Gene). HISAT2 (http://www.ccb.jhu.edu/software/hisat) was used to map clean reads to the genomic sequence, and the results showed that the total read numbers of each sample were around 23.8 M, and the coverage was 94.8–96.2%.
Next, RSEM (https://deweylab.biostat.wisc.edu/rsem/rsem-calculate-expression.html) was used to calculate the gene expression level of each sample and DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) was used for differential expression analysis, of which differentially expressed genes (DEGs) were expressed as a Q value (adjusted P value) ≤0.05. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed according to the Dr. Tom approach, an in-house customized data mining system of the Shenzhen Genomics Institute (BGI-Huada Gene).
Statistical analysis
All experiments were repeated at least 3 times. Data were expressed as the mean ± standard deviation (SD). Statistical analysis was performed using SPSS 20.0 software (IBM Corp., Armonk, NY, USA). Differences among groups were analyzed using analysis of variance (ANOVA) or Student’s t-test. A P value <0.05 was considered statistically significant.
Results
MEF2D is overexpressed in B-ALL and correlates with the disease progression of ALL
To illustrate the differential expression of MEF2D in B-ALL patients and healthy donors, two datasets (GSE13159, GSE48558) from the Gene Expression Omnibus (GEO) database were utilized to confirm that MEF2D mRNA expression was significantly upregulated in B-ALL patients compared to the healthy donors (Figure 1A,1B). Additionally, to illustrate the differential expression of MEF2D in B-ALL and normal B cells, we analyzed GSE34861 containing 28 pro-B-ALL, 153 pre-B-ALL, and 3 normal pre-B cells, suggesting that MEF2D expression was higher in pro-B-ALL and pre-B-ALL than in normal pre-B cells (Figure S1A). Moreover, MEF2D mRNA expression was higher in pre-B-ALL than in CD10+CD19+B cells in GSE34670 (Figure S1B). Next, we evaluated MEF2D mRNA expression in bone marrow samples from B-ALL patients and healthy donors by qRT-PCR. The results suggested that MEF2D mRNA expression was elevated in B-ALL patients compared with healthy donors (Figure 1C).
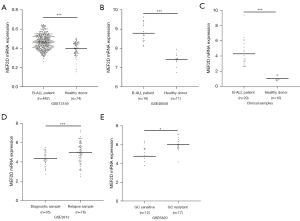
To further validate the correlation between MEF2D expression and the disease progression of ALL patients, we analyzed the GSE3912 dataset containing 35 initial diagnosis patients and 78 relapsed patients, indicating that MEF2D expression was higher in relapsed patients compared to initial diagnosis patients (Figure 1D). Moreover, MEF2D mRNA expression was higher in glucocorticoid-resistant patients than in glucocorticoid-sensitive patients in GSE5820 (Figure 1E). These results demonstrated that MEF2D was overexpressed in B-ALL patients and correlated with ALL disease progression.
Knockdown of MEF2D suppresses cell viability, induces cell apoptosis, and blockades cell cycle progression in B-ALL cells
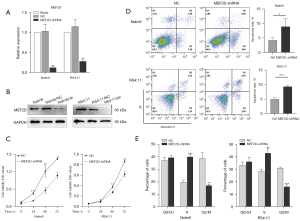
To explore the potential role of MEF2D in the progression of B-ALL, we transduced Nalm6 and RS4;11 cells with MEF2D-shRNA or its NC lentivirus vector, respectively. As illustrated in Figure 2A,2B, MEF2D expression was decreased at both the mRNA and protein levels in MEF2D-shRNA-transduced Nalm6 and RS4;11 cells, as compared to the NC cells. Our results showed that MEF2D knockdown significantly inhibited cell viability (Figure 2C) and promoted cell apoptosis (Figure 2D) compared to the NC cells. FCM analysis demonstrated that MEF2D knockdown blockaded cell cycle progression from the S phase to the G2/M phase (Figure 2E). In conclusion, these results suggested that MEF2D knockdown suppressed cell viability, induced cell apoptosis, and blockaded cell cycle progression in B-ALL.
Knockdown of MEF2D enhances drug sensitivity in B-ALL cells
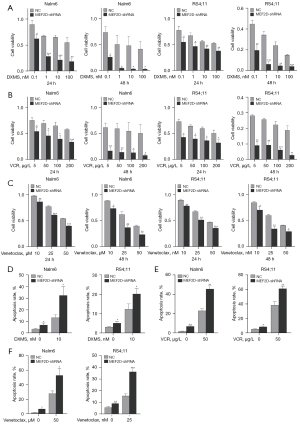
The current standard chemotherapy regimen for the treatment of B-ALL includes DXMS and VCR. Additionally, venetoclax, a selective BCL-2 inhibitor, is also considered a promising drug for B-ALL treatment (17,18). Therefore, DXMS, VCR, and venetoclax were used in this study to investigate the effect of MEF2D on drug sensitivity in B-ALL cells. The cell viability and cell apoptosis of MEF2D-shRNA and NC cells after diverse drug treatments were evaluated by the CCK-8 assay and FCM, respectively. As shown in Figure 3A-3C, MEF2D knockdown enhanced the drug sensitivity of human B-ALL cells to DXMS, VCR, and venetoclax. Furthermore, we demonstrated that MEF2D knockdown also enhanced cell apoptosis induced by DXMS, VCR, and venetoclax via FCM (Figure 3D-3F). Taken together, these results indicated that MEF2D knockdown enhanced the drug sensitivity in B-ALL.
Knockdown of MEF2D inhibits B-ALL cell growth in vivo
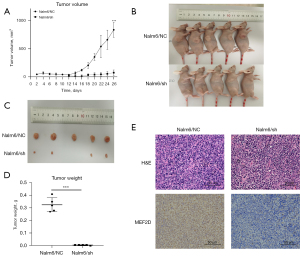
Previous results confirmed that MEF2D knockdown inhibited the cell viability of B-ALL in vitro. To further explore the effect of MEF2D on tumor growth in vivo, we established a B-ALL xenograft mouse model by injecting Nalm6/NC or Nalm6/sh cells into Balb/c nude mice, respectively. The tumor volume was measured every 2 days. As illustrated, mice injected with Nalm6/sh cells exhibited delayed tumorigenesis compared with the Nalm6/NC mice (Figure 4A). The mice were then euthanized, and tumors were removed and measured on day 26. As expected, the tumor volumes were significantly suppressed in mice infused with Nalm6/sh compared to those infused with Nalm6/NC (Figure 4B-4D). IHC staining was then performed to confirm that MEF2D expression was lower in the tumors of Nalm6/sh mice compared to those in Nalm6/NC mice (Figure 4E). As above, these results demonstrated that MEF2D played an important role in the tumorigenicity of B-ALL in vivo.
Knockdown of MEF2D downregulates the expression of the PI3K-AKT signaling pathway
To elucidate the mechanisms through which MEF2D regulates B-ALL cells, we performed RNA sequencing on Nalm6/NC and Nalm6/sh cells. A total of 16,860 genes were detected, and 15,083 genes were shared between the NC cells and MEF2D knockdown cells (Figure 5A). The results indicated that there were 842 DEGs, of which 521 genes were upregulated and 321 genes were downregulated (Figure 5B).
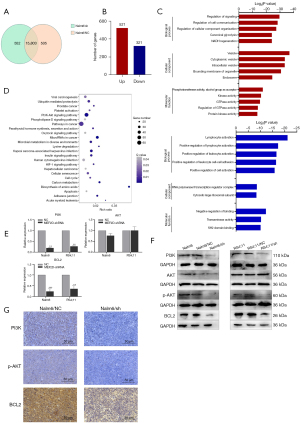
The GO classification of upregulated genes indicated that these genes were mainly distributed in biological processes (BPs) such as the regulation of signaling and cell communication, cellular components (CCs) such as the vesicle and endosome, and molecular functions (MFs) such as kinase and phosphotransferase activities. Meanwhile, GO classification of the downregulated genes indicated that these genes were mainly distributed in BPs such as lymphocyte activation and the positive regulation of leukocyte cell-cell adhesion, CCs such as the RNA polymerase II regulator complex and the cytosolic large ribosomal subunit, and MFs such as negative regulation of binding and transaminase activity (Figure 5C).
Based on the RNA sequencing results, KEGG analysis of the DEGs was performed to identify the signaling pathways in which the DEGs were mainly involved, among which the PI3K-AKT signaling pathway and pathways in cancer involved a majority of DEGs (Figure 5D). We conducted qRT-PCR and western blotting to validate the key genes expression of the PI3K-AKT signaling pathway between the MEF2D-shRNA and NC cells, suggesting that MEF2D knockdown significantly downregulated the expression of some key genes in the PI3K-AKT signaling pathway, such as PI3K, p-AKT, and BCL2 (Figure 5E,5F).
Also, the IHC staining results showed that the expressions of PI3K, p-AKT, and BCL2 in the tumor tissue of the Nalm6/sh group were lower than those of the Nalm6/NC group (Figure 5G). These results indicated that MEF2D knockdown downregulated the expression of the PI3K-AKT signaling pathway in B-ALL.
Discussion
In mammals, MEF2D is the most commonly expressed of the 4 homologs, being abundantly expressed in almost all tissues and cell types. This wide-ranging expression pattern reveals the vital role of MEF2D in modulating various BPs, including mitochondrial activity, cell viability, cell apoptosis, cell migration, and cell cycle progression (9). In normal hemopoiesis, both MEF2C and MEF2D are activated and involved in early B-cell development (19); however, MEF2D knockout does not cause significant B-cell development defects (19,20). Besides, neither the development of T cells nor myeloid cells appear to be affected by MEF2D deletion (20,21). During development, MEF2D-null mice are viable and appear normal (22). Overall, previous studies have indicated that the loss of MEF2D alone does not have a major impact on development.
Over the past several years, recurrent fusions between MEF2D and distinct partner genes have been identified in B-ALL, with frequencies of 2–7% and 2–4% in adult and pediatric patients, respectively (23-25). A total of 9 genes (BCL9, SS18, FOXJ2, CSF1R, DAZAP1, STAT6, HNRNPUL1, HNRNPH1, and HNRNPM) have been reported as fusion partners of MEF2D (26). Patients with MEF2D fusions have formed a unique group with a pre-B cell immunophenotype, the arrest of B-cell differentiation, and a poor clinical outcome. Thus, MEF2D fusion has been considered an unfavorable subtype (15,26,27). Rearrangements result in enhanced MEF2D transcriptional activity. Also, the strictly conserved MADS-box domain and the extreme similarity in gene expression with different MEF2D fusions suggest that the leukemogenesis may be driven by the MEF2D TF itself (24,26,28). These findings indicate that MEF2D is a potential therapeutic target in B-ALL.
Nevertheless, prior to this study, the function and mechanism of MEF2D in B-ALL has remained unclear. In this study, we found that MEF2D expression was upregulated in B-ALL patients compared with healthy controls and normal B cells, which made it an attractive target in B-ALL. To verify the effect of MEF2D in B-ALL, we constructed MEF2D-knockdown B-ALL cell lines via lentivirus transfection. The results indicated that MEF2D knockdown in B-ALL significantly inhibited cell viability, induced cell apoptosis, and blocked the cell cycle in the G2/M phase in vitro. Moreover, the knockdown of MEF2D also enhanced the sensitivity of B-ALL to first-line chemotherapeutics and a novel BCL2 inhibitor. To further confirm the effect of MEF2D in B-ALL, we established the B-ALL xenograft mouse model and confirmed that MEF2D knockdown inhibited tumor growth in vivo. In conclusion, our data illustrated that MEF2D expression was crucial in B-ALL tumor progression in vivo and in vitro.
The PI3K-AKT signaling pathway is involved in a variety of physiological processes, such as cell growth, cell proliferation, cell survival, cell migration, and cell apoptosis (29). Hyperactivation of the PI3K signaling cascade is one of the most common occurrences in human carcinoma. Multiple studies have demonstrated that the PI3K-AKT signaling pathway is overexpressed in a variety of human solid tumors and hematological malignancies (30-32). Moreover, it has been universally reported that the PI3K-AKT signaling pathway is hyperactive in ALL, promoting the survival of leukemic stem cells as well as the development of the leukemic bone marrow microenvironment and drug resistance (33-35). In addition, studies have confirmed that dysregulated expression of apoptosis TFs can promote the incidence of leukemia, and the overexpression of BCL2 is associated with leukemia drug resistance (36-39).
This study explored the mechanism by which MEF2D regulates the tumor progression of B-ALL via RNA sequencing. The results showed that the DEGs were significantly enriched in the PI3K-AKT signaling pathway. Furthermore, we verified that MEF2D knockdown downregulated the expression of key regulators in the PI3K-AKT pathway via qRT-PCR, western blotting, and IHC staining. In addition, our data demonstrated that MEF2D knockdown downregulated BCL2 expression at both the mRNA and protein levels. In summary, MEF2D may act as a key factor to regulate the development and progression of B-ALL, which may be related to the PI3K-AKT signaling pathway.
In summary, we sought to confirm the role of MEF2D in B-ALL tumorigenesis. We found that MEF2D knockdown could inhibit cell viability and tumor growth, induce cell apoptosis, blockade cell cycle progression, and increase drug sensitivity in B-ALL cells. Furthermore, MEF2D knockdown also downregulated the expression of the PI3K-AKT signaling pathway. Our finding suggested that MEF2D is a promising molecular target for B-ALL. However, further studies are required to elucidate the underlying molecular mechanisms through which MEF2D regulates the PI3K-AKT signaling pathway. In addition, more comprehensive investigations to fully validate the clinical value of MEF2D in B-ALL should be performed in future studies with larger sample sizes and longer follow-up periods.
Conclusions
In conclusion, our research demonstrated that MEF2D was markedly expressed in B-ALL, and the knockdown of MEF2D inhibited cancer progression of B-ALL in vitro and in vivo, which may be related to the downregulation of the PI3K-AKT signaling pathway. The results of this study suggested that MEF2D plays a vital role in the process of tumorigenesis in B-ALL and may be a potential novel target for B-ALL therapy.
Acknowledgments
Funding: This work was supported by the Frontier Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (No. 2018GZR110105014) and the Science and Technology Planning Project of Guangdong Province, China (No. 2018B030311042).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1778/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1778/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1778/coif). All authors report that this work was supported by the Frontier Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (No. 2018GZR110105014) and the Science and Technology Planning Project of Guangdong Province, China (No. 2018B030311042). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by the Ethics Committee of the Zhujiang Hospital of Southern Medical University (No. 2022-KY-129). Informed consent was provided by all individual participants. The animal experiments were performed under a project license (No. LAEC-2020-117) granted by the Animal Ethics Committee of Zhujiang Hospital of Southern Medical University, in compliance with Zhujiang Hospital’s guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Kline KAF, Kallen ME, Duong VH, et al. Acute Lymphoblastic Leukemia and Acute Lymphoblastic Lymphoma: Same Disease Spectrum but Two Distinct Diagnoses. Curr Hematol Malig Rep 2021;16:384-93. [Crossref] [PubMed]
- Inaba H, Pui CH. Advances in the Diagnosis and Treatment of Pediatric Acute Lymphoblastic Leukemia. J Clin Med 2021;10:1926. [Crossref] [PubMed]
- Neaga A, Jimbu L, Mesaros O, et al. Why Do Children with Acute Lymphoblastic Leukemia Fare Better Than Adults? Cancers (Basel) 2021;13:3886. [Crossref] [PubMed]
- Sasaki K, Jabbour E, Short NJ, et al. Acute lymphoblastic leukemia: A population-based study of outcome in the United States based on the surveillance, epidemiology, and end results (SEER) database, 1980-2017. Am J Hematol 2021;96:650-8. [Crossref] [PubMed]
- Advani AS. Novel strategies in the treatment of acute lymphoblastic leukaemia. Lancet Haematol 2022;9:e240-1. [Crossref] [PubMed]
- Brown PA, Shah B, Advani A, et al. Acute Lymphoblastic Leukemia, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2021;19:1079-109. [Crossref] [PubMed]
- Jen EY, Xu Q, Schetter A, et al. FDA Approval: Blinatumomab for Patients with B-cell Precursor Acute Lymphoblastic Leukemia in Morphologic Remission with Minimal Residual Disease. Clin Cancer Res 2019;25:473-7. [Crossref] [PubMed]
- Madugula K, Mulherkar R, Khan ZK, et al. MEF-2 isoforms' (A-D) roles in development and tumorigenesis. Oncotarget 2019;10:2755-87. [Crossref] [PubMed]
- Shen P, Yu Y, Yan Y, et al. LncRNA CASC15 regulates breast cancer cell stemness via the miR-654-5p/MEF2D axis. J Biochem Mol Toxicol 2022;36:e23023. [Crossref] [PubMed]
- Xiang J, Zhang N, Sun H, et al. Disruption of SIRT7 Increases the Efficacy of Checkpoint Inhibitor via MEF2D Regulation of Programmed Cell Death 1 Ligand 1 in Hepatocellular Carcinoma Cells. Gastroenterology 2020;158:664-678.e24. [Crossref] [PubMed]
- Xu J, Li Y, Li Z, et al. Acidic Tumor Microenvironment Promotes Pancreatic Cancer through miR-451a/MEF2D Axis. J Oncol 2022;2022:3966386. [Crossref] [PubMed]
- Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet 2002;32:166-74. [Crossref] [PubMed]
- Vega-García N, Malatesta R, Estella C, et al. Paediatric patients with acute leukaemia and KMT2A (MLL) rearrangement show a distinctive expression pattern of histone deacetylases. Br J Haematol 2018;182:542-53. [Crossref] [PubMed]
- Ohki K, Kiyokawa N, Saito Y, et al. Clinical and molecular characteristics of MEF2D fusion-positive B-cell precursor acute lymphoblastic leukemia in childhood, including a novel translocation resulting in MEF2D-HNRNPH1 gene fusion. Haematologica 2019;104:128-37. [Crossref] [PubMed]
- Cao Y, Tang S, Nie X, et al. Decreased miR-214-3p activates NF-κB pathway and aggravates osteoarthritis progression. EBioMedicine 2021;65:103283. [Crossref] [PubMed]
- Yang W, Cai J, Shen S, et al. Pulse therapy with vincristine and dexamethasone for childhood acute lymphoblastic leukaemia (CCCG-ALL-2015): an open-label, multicentre, randomised, phase 3, non-inferiority trial. Lancet Oncol 2021;22:1322-32. Erratum in: Lancet Oncol 2021;22:e389. [Crossref] [PubMed]
- Smith WM, Reed DR. Targeting Apoptosis in ALL. Curr Hematol Malig Rep 2022;17:53-60. [Crossref] [PubMed]
- Herglotz J, Unrau L, Hauschildt F, et al. Essential control of early B-cell development by Mef2 transcription factors. Blood 2016;127:572-81. [Crossref] [PubMed]
- Pattison MJ, Naik RJ, Reyskens KMSE, et al. Loss of Mef2D function enhances TLR induced IL-10 production in macrophages. Biosci Rep 2020;40:BSR20201859. [Crossref] [PubMed]
- Pingul BY, Huang H, Chen Q, et al. Dissection of the MEF2D-IRF8 transcriptional circuit dependency in acute myeloid leukemia. iScience 2022;25:105139. [Crossref] [PubMed]
- Kim Y, Phan D, van Rooij E, et al. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest 2008;118:124-32. [Crossref] [PubMed]
- Gu Z, Churchman M, Roberts K, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun 2016;7:13331. [Crossref] [PubMed]
- Li J, Dai Y, Wu L, et al. Emerging molecular subtypes and therapeutic targets in B-cell precursor acute lymphoblastic leukemia. Front Med 2021;15:347-71. [Crossref] [PubMed]
- Paietta E, Roberts KG, Wang V, et al. Molecular classification improves risk assessment in adult BCR-ABL1-negative B-ALL. Blood 2021;138:948-58. [Crossref] [PubMed]
- Zhang M, Zhang H, Li Z, et al. Functional, structural, and molecular characterizations of the leukemogenic driver MEF2D-HNRNPUL1 fusion. Blood 2022;140:1390-407. [Crossref] [PubMed]
- Leo IR, Aswad L, Stahl M, et al. Integrative multi-omics and drug response profiling of childhood acute lymphoblastic leukemia cell lines. Nat Commun 2022;13:1691. [Crossref] [PubMed]
- Sadras T, Müschen M. MEF2D Fusions Drive Oncogenic Pre-BCR Signaling in B-ALL. Blood Cancer Discov 2020;1:18-20. [Crossref] [PubMed]
- Vasan N, Cantley LC. At a crossroads: how to translate the roles of PI3K in oncogenic and metabolic signalling into improvements in cancer therapy. Nat Rev Clin Oncol 2022;19:471-85. [Crossref] [PubMed]
- Sanaei MJ, Baghery Saghchy Khorasani A, Pourbagheri-Sigaroodi A, et al. The PI3K/Akt/mTOR axis in colorectal cancer: Oncogenic alterations, non-coding RNAs, therapeutic opportunities, and the emerging role of nanoparticles. J Cell Physiol 2022;237:1720-52. [Crossref] [PubMed]
- Meng D, He W, Zhang Y, et al. Development of PI3K inhibitors: Advances in clinical trials and new strategies Pharmacol Res 2021;173:105900. (Review). [Crossref] [PubMed]
- He Y, Sun MM, Zhang GG, et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther 2021;6:425. [Crossref] [PubMed]
- Kim HN, Ogana H, Sanchez V, et al. PI3K Targeting in Non-solid Cancer. Curr Top Microbiol Immunol 2022;436:393-407. [Crossref] [PubMed]
- Chen Q, Lai Q, Jiang Y, et al. Anlotinib exerts potent antileukemic activities in Ph chromosome negative and positive B-cell acute lymphoblastic leukemia via perturbation of PI3K/AKT/mTOR pathway. Transl Oncol 2022;25:101516. [Crossref] [PubMed]
- Ehm P, Grottke A, Bettin B, et al. Investigation of the function of the PI3-Kinase/AKT signaling pathway for leukemogenesis and therapy of acute childhood lymphoblastic leukemia (ALL). Cell Signal 2022;93:110301. [Crossref] [PubMed]
- Aumann S, Shaulov A, Haran A, et al. The Emerging Role of Venetoclax-Based Treatments in Acute Lymphoblastic Leukemia. Int J Mol Sci 2022;23:10957. [Crossref] [PubMed]
- Richter A, Lange S, Holz C, et al. Effective tumor cell abrogation via Venetoclax-mediated BCL-2 inhibition in KMT2A-rearranged acute B-lymphoblastic leukemia. Cell Death Discov 2022;8:302. [Crossref] [PubMed]
- Valentin R, Grabow S, Davids MS. The rise of apoptosis: targeting apoptosis in hematologic malignancies. Blood 2018;132:1248-64. [Crossref] [PubMed]
- Gibson A, Trabal A, McCall D, et al. Venetoclax for Children and Adolescents with Acute Lymphoblastic Leukemia and Lymphoblastic Lymphoma. Cancers (Basel) 2021;14:150. [Crossref] [PubMed]


