
A five-gene expression signature of centromeric proteins with prognostic value in lung adenocarcinoma
Highlight box
Key findings
• In this study, we assessed the prognostic value of the centromere proteins (CENP) family proteins and constructed a risk model involving CENPF, CENPU, CENPM, CENPH, and CENPW in lung adenocarcinoma (LUAD).
What is known and what is new?
• In previous studies, the CENP family proteins were generally found to be hyperexpressed in various types of cancers and associated with the clinical characteristics and outcomes based on their roles in cell mitosis. At present, there is no comprehensive screening and systemic evaluation of the CENP protein family in malignant tumors, especially LUAD.
• In this study, differentially expressed CENPs at the transcriptional level were screened, and their prognostic value and correlations with clinicopathological parameters, genetic alteration, and the coexpression pattern were revealed. Moreover, a risk model involving CENPs was constructed using the least absolute shrinkage and selection operator (LASSO).
What are the implications, and what should change now?
• This study provided a risk model for prognostic assessment and identified potential therapeutic targets in LUAD.
• Additional clinical data are needed to validate the model. Furthermore, the interaction mechanism between CENPs and downstream molecules should be further explored.
Introduction
Lung cancer remains the leading cause of cancer-related death worldwide, with significant morbidity and mortality rates (1). Lung adenocarcinoma (LUAD) is the most common lung cancer subtype, with an average 5-year survival rate of 15% (2,3), and most patients are diagnosed at an advanced stage (4,5). Thus, the discovery of reliable biomarkers is critical for determining the prognosis of LUAD. Chromosomal instability (CIN) is a marker of cancer in almost 90% of human tumors (6-8), and abnormal expression of the centromere protein (CENP) family is closely related to CIN (9-11).
CENP is a large protein family with more than 20 members, which are mainly involved in the constitutive centromere-associated network (CCAN) (12-14), a group of 16 CENP family proteins positioned at the centromere throughout the cell cycle (14). CCAN forms the centromere base connecting the centromere and microtubule. In the CCAN, CENP family proteins are divided into several functional groups: CENPC, CENP-H/I/K, CENP-L/M/N, CENP-O/P/Q/R/U, and CENP-T/W/S/X (15). For instance, CENPA, also known as histone H3-like centromeric protein A, which is replicated during the S-phase, is involved in the formation of the centromeric nucleosome structure and is essential for the localization of all known kinetochore components (14). CENPA interacts with CENP-C/N and participates in mitotic progression and chromosome segregation (12). Accumulating at the G2 phase of the cell cycle, CENPE acts as kinesin to link kinetochores to the releasing microtubule plus-end and interacts with mitogen-activated protein (MAP) kinases and extracellular signal-regulated kinases 1 and 2 (ERK1 and ERK2) (16,17). It has been found that the expression of CENP family proteins is upregulated in several cancers and is related to the advanced cancer characteristics of patients, including clinical stage, grade, and metastasis (11,16-19).
Several CENP proteins are reported to be associated with multiple common cancers, including hepatocellular carcinoma, breast cancer, gastric cancer, and lung cancer; however, there is currently a lack of research on the CENP protein family (11,16,17,20-22). The reduced expression of CENPE or overexpression of CENPH contributes to tumor progression, and these may be novel prognostic biomarkers in human hepatocellular carcinoma (20,22). Moreover, the high expression of CENPH is related to poor prognosis in patients with gastric cancer (21). In lung cancer, CENPA upregulation is associated with poor prognosis and may serve as a potential therapeutic target for patients with LUAD (23), CENPE regulated by FOXM1 promotes LUAD proliferation (17), and CENPH has been reported to be a prognostic biomarker for patients with non-small cell lung cancer (NSCLC) (24). However, no comprehensive screening of the CENP protein family in malignant tumors has been conducted thus far. Moreover, the existing research is mostly based on small sample sizes and does not consider the interaction between CENP family members.
In the present study, bioinformatics analysis based on several large online databases was performed to explore the relationship between CENPs and clinicopathological parameters and their prognostic value in LUAD (Figure 1). We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2166/rc).
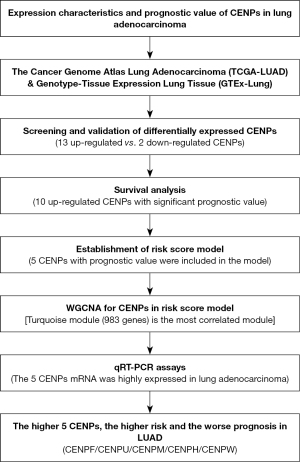
Methods
Gene databases
We integrated the RNA-sequencing (RNA-seq) data of 526 LUAD tissues and 59 normal lung tissues from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov) and 288 normal lung tissues from the Genotype-Tissue Expression (GTEx) (25) database (https://gtexportal.org/). These data were used to evaluate the CENP expression differences between LUAD and normal lung tissues.
Selection of differential genes
We analyzed most members of the CENP protein family. To screen CENPs that are differentially expressed in LUAD and normal lung tissues, we used the “limma” package in Bioconductor (Bioconductor—Open Source Software for Bioinformatics Copyright 2017) and R version 3.2.5 (The R Project for Statistical Computing) packages. to analyze the expression of CENP family proteins in 526 tumor tissues and 347 normal lung tissues. The screening threshold was |log fold change (FC)| >1.0 and adjusted P<0.05. A heat map and violin plot were used to demonstrate the gene expression levels.
Survival analysis of differential CENPs
The Kaplan-Meier method was used to analyze the survival of 526 patients with LUAD to explore the prognostic value of differentially expressed CENPs in patients with LUAD. Moreover, the “corrplot” package in R was used to visualize the correlation among CENPs.
Cox proportional hazards models
We obtained the survival time and status of 526 patients with LUAD from TCGA database and then constructed a risk model of CENPs with differential expression using the least absolute shrinkage and selection operator (LASSO) Cox regression algorithm. Kaplan-Meier survival analysis, receiver operating characteristic (ROC) curve, and univariate and multivariate Cox regression analysis were performed to evaluate the accuracy of the risk model.
Gene coexpression network analysis
The coexpression network of differentially expressed CENPs was constructed by weighted gene correlation network analysis (WGCNA) in R (The R Foundation of Statistical Computing), and the corresponding hierarchical clustering and gene modules were generated to screen out gene modules that were closely related to the clinical features and differential expression of the CENP protein family with the prognostic value for LUAD.
RNA extraction and quantitative real time polymerase chain reaction (qRT-PCR)
We used the RNAprep FastPure Tissue & Cell Kit (Tsingke Biotechnology, Beijing, China) and ABScript III Reverse Transcriptase (ABclonal, Wuhan, China) to extract messenger RNA (mRNA) from LUAD and normal tissue and performed reverse transcription according to the manufacturer’s protocol. Finally, qRT-PCR experiments were carried out using ABScript II One-Step SYBR Green RT-qPCR Kit (ABclonal). The primer sequences were as follows: CENPW: 5'-GAT GGA ACT GGC TGA GAC ACT AAC C-3' (forward) and 5'-AAG ACT CTT GCT TGA TGC TGA GGT G-3' (reverse); CENPM: 5'-ACA GCA AAT ACA GTC TCC AGA A-3' (forward) and 5'-GAA ACA CAC CTT CCC CAA GAA-3' (reverse); CENPU: 5'-GAA AAG AAA AGG CAG CGT ATG A-3' (forward) and 5'-AAT ATG CTG CAT TCC TAA GGG A-3' (reverse); CENPF: 5'-TAC AAC GAG AGA GTA AGA ACG C-3' (forward) and 5'-CTA CCT CCA CTG ACT TAC TGT C-3' (reverse); CENPH: 5'-TTC CAG AAC CTT ATT TTG GGG A-3' (forward) and 5'-CTT CTC AAG CTG CAG AAC AAT T-3' (reverse).
Patients and tissue samples
A total of 5 patients LUAD underwent surgical resection at the Tongji Hospital of Huazhong University of Science and Technology, Tongji Medical College (Wuhan, China). Snap-frozen tissues from these patients were collected at Tongji Hospital in 2018. The 5 patients were histologically diagnosed with primary LUAD. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). It was approved by the Research Ethics Committee, Tongji Medical College, Huazhong University of Science and Technology (No. 20180403). Informed consent was waived due to the retrospective nature of this study.
Statistical analysis
Following a normality check, the Student’s t-test was used to evaluate normally distributed data, and Mann-Whitney tests were used to analyze nonnormally distributed data. The correlation between CENPs was examined using the Pearson test, while the correlation between these DEGs and clinicopathologic characteristics was examined with Kendall rank correlation coefficient testing. The prognostic significance of CENPs was assessed using Kaplan-Meier survival analysis and Cox multivariate regression analysis. Differences were considered to be statistically significant at P<0.05.
Results
Differential expression analyses of CENP family members in LUAD
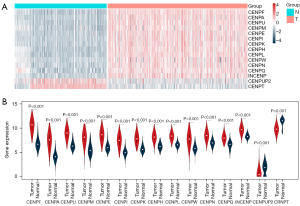
A total of 15 differentially expressed CENP family members were screened out (Table 1), and 13 CENP family members were upregulated compared with the normal tissues: CENPF, CENPA, CENPU, CENPM, CENPE, CENPI, CENPK, CENPH, CENPL, CENPW, CENPN, CENPQ, and INCENP (Figure 2A). Two CENP family members were downregulated, namely CENPUP2 (logFC –1.028; P<0.001) and CENPT (logFC –1.514; P<0.001) (Figure 2A). The violin plots showed that CENPF (logFC 3.720; P<0.001) were significantly upregulated and that CENPUP2 (logFC –1.02; P<0.001) were significantly downregulated in tumor tissues (Figure 2B).
Table 1
| CENPs | LogFC | AveExpr | t | P | Adj.P | β | Change |
|---|---|---|---|---|---|---|---|
| CENPA | 3.247537077 | 6.117932933 | 34.22669389 | 2.33e−163 | 9.47e−162 | 362.8805908 | Up |
| CENPF | 3.720029395 | 9.063292383 | 41.98118255 | 1.89e−211 | 2.29e−209 | 473.5150655 | Up |
| CENPE | 2.458642295 | 7.557335664 | 31.86512482 | 2.60e−148 | 8.00e−147 | 328.2642046 | Up |
| CENPB | 0.174938294 | 11.7181103 | 4.425952599 | 1.08e−05 | 1.90e−05 | 1.304591913 | Stable |
| INCENP | 1.370221774 | 9.160947297 | 27.47616633 | 3.51e−120 | 6.43e−119 | 263.5573823 | Up |
| CENPC | −0.04437037 | 9.519976219 | −1.36183773 | 0.1736013 | 0.2034584 | −7.46063137 | Stable |
| CENPJ | 0.09992807 | 8.58660911 | 2.284476046 | 0.0225834 | 0.0307698 | −5.78425074 | Stable |
| CENPH | 1.982387332 | 7.431167486 | 34.49783924 | 4.46e−165 | 1.87e−163 | 366.8351839 | Up |
| CENPU | 2.547406909 | 7.965051181 | 36.17931184 | 1.10e−175 | 5.75e−174 | 391.243498 | Up |
| CENPW | 1.796088492 | 7.014250978 | 22.57557371 | 3.32e−89 | 3.50e−88 | 192.3280442 | Up |
| CENPM | 2.518778578 | 7.036015964 | 28.81203931 | 9.67e−129 | 2.05e−127 | 283.2474704 | Up |
| CENPO | 0.325312428 | 8.74232919 | 7.0502545 | 3.64e−12 | 8.42e−12 | 15.79720682 | Stable |
| CENPT | −1.51423458 | 10.37256876 | −26.5606414 | 2.50e−114 | 4.11e−113 | 250.0991236 | Down |
| CENPI | 2.382413306 | 6.427262231 | 31.03463241 | 5.39e−143 | 1.49e−141 | 316.0347634 | Up |
| CENPN | 1.540384156 | 7.87364105 | 25.98125103 | 1.23e−110 | 1.90e−109 | 241.6059193 | Up |
| CENPK | 2.132952807 | 6.968600679 | 31.57624334 | 1.84e−146 | 5.44e−145 | 324.0126868 | Up |
| CENPV | 0.135675099 | 8.289197752 | 2.122279385 | 0.034096 | 0.0444279 | −6.13987356 | Stable |
| CENPP | −0.29374296 | 7.494897026 | −6.92150784 | 8.66e−12 | 1.98e−11 | 14.9450106 | Stable |
| CENPQ | 1.434607439 | 7.449989654 | 25.70371121 | 7.18e−109 | 1.07e−107 | 237.545656 | Up |
| CENPL | 1.826436692 | 7.650182282 | 37.11307898 | 1.55e−181 | 9.36e−180 | 404.7019582 | Up |
| CENPCP1 | 0.408609084 | 1.445567364 | 5.519305999 | 4.49e−08 | 8.91e−08 | 6.592921301 | Stable |
| CENPUP2 | −1.02813774 | 1.176967002 | −13.6614034 | 1.22e−38 | 5.34e−38 | 76.21181011 | Down |
| CENPIP1 | 0.085278812 | 0.058108903 | 2.886643811 | 0.00399 | 0.0058634 | −4.23834699 | Stable |
| CENPUP1 | 0.006482708 | 0.021025974 | 0.659995781 | 0.5094311 | 0.5467224 | −8.16965633 | Stable |
CENPs, centromere proteins; FC, fold change; AveExpr, average expression; Adj., adjusted.
Prognostic values of CENP family members in patients with LUAD
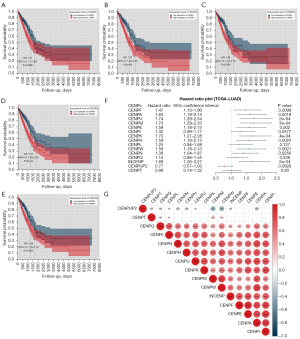
For differentially expressed CENP family members, we performed survival analysis using the “survival” and “survcomp” R packages. Prognostic information was obtained from 513 patients with LUAD in TCGA database. Among the 15 differentially expressed CENP family members, 10 proteins had a significant prognostic value, including CENPF, CENPA, CENPU, CENPM, CENPE, CENPK, CENPH, CENPW, CENPN, and INCENP. High expression of these proteins predicted a shorter overall survival (OS) (Figure 3A-3F). We then assessed the connection between the mRNA levels of 15 CENP family members and found that CENPF, CENPA, CENPU, CENPM, CENPE, CENPK, CENPH, CENPW, CENPN, and INCENP were well correlated with each other as prognostic factors (Figure 3G).
Construction of the risk model and its relationship with clinicopathological features and prognosis
To evaluate the relationship between CENP family members and clinical outcomes, we established a risk assessment model for 10 genes in TCGA LUAD data set using the LASSO Cox regression algorithm (Figure 4A,4B). The LASSO algorithm screened genes that were closely related to prognosis, including CENPF, CENPU, CENPM, CENPH, and CENPW, and then constructed the risk prediction model. We divided TCGA LUAD data set into high-risk (n=248) and low-risk (n=265) groups according to the median risk score to evaluate the correlation between the risk model and clinical characteristics (Table 2). The results showed that the risk model was related to the T stage (P<0.001) and M stage (P<0.05) (Figure 4C). The Kaplan-Meier curve showed that the prognosis of the high-risk group was poor [hazard ratio (HR): 1.75, 95% confidence interval (CI): 1.30–2.35; P=2e−04]. At the same time, the area under the ROC curve (AUC) values for the 1-, 3-, and 5-year survival were 0.63, 0.62, and 0.6, respectively, indicating that the diagnostic value of risk model was not strong (Figure 4D,4E).
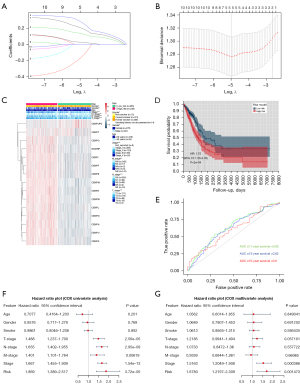
Table 2
| Characteristics | No. of patients (%) |
|---|---|
| Age (years) | |
| <50 | 35 (6.82) |
| ≥50 | 478 (93.18) |
| Gender | |
| Male | 237 (46.20) |
| Female | 276 (53.80) |
| Smoking | |
| Nonsmoker | 72 (14.04) |
| Current smoker | 121 (23.59) |
| Former smoker | 306 (59.65) |
| Smoking history not documented | 14 (2.73) |
| Pathological tumor (T) statusa | |
| T1 | 171 (33.33) |
| T2 | 275 (53.61) |
| T3 | 46 (8.97) |
| T4 | 18 (3.51) |
| TX | 3 (0.58) |
| Pathological node (N) statusa | |
| N0 | 335 (65.30) |
| N1 | 94 (18.32) |
| N2 | 69 (13.45) |
| N3 | 2 (0.39) |
| NX | 13 (2.53) |
| Pathological metastasis (M) statusa | |
| M0 | 342 (66.67) |
| M1 | 17 (3.31) |
| M1a | 2 (0.39) |
| M1b | 5 (0.97) |
| MX | 147 (28.65) |
| Clinical stagea | |
| I | 280 (54.58) |
| II | 120 (23.39) |
| III | 80 (15.59) |
| IV | 25 (4.87) |
| Risk score | |
| Low | 265 (51.66) |
| High | 248 (48.34) |
a, pathological tumor (T) status, pathological node (N) status, and clinical stage are from the eighth edition of Union for International Cancer Control (UICC)/American Joint Committee on Cancer (AJCC) lung cancer stage classification [2017]. TCGA-LUAD, The Cancer Genome Atlas-lung adenocarcinoma.
We then performed Cox regression to analyze the LUAD data of TCGA by univariate and multivariate analysis. Univariate Cox regression analysis found that stage (HR: 1.667, 95% CI: 1.455–1.909; P=1.54e−13), T stage (HR: 1.488, 95% CI: 1.237–1.79; P=2.56e−05), N stage (HR: 1.655, 95% CI: 1.402–1.955; P=2.95e−09), M stage (HR: 1.401, 95% CI: 1.101–1.784; P=0.00619), and risk score (HR: 1.869, 95% CI: 1.389–2.517; P=3.72e−05) were correlated with prognosis. Multivariate Cox regression analysis showed that stage (HR: 1.5163, 95% CI: 1.2049–1.908; P=0.000386) and risk score (HR: 1.678, 95% CI: 1.2197–2.309; P=0.001473) were correlated with prognosis (Figure 4F,4G).
To verify the expression levels of CENPM, CENPW, CENPU, CENPF, and CENPH in LUAD tissues, we collected tumor specimens from 5 patients with LUAD and paired normal tissues for qRT-PCR. We observed that the mRNA expression levels of the above 5 genes in tumor tissues were higher than those in corresponding normal tissues. The mRNA expression level of CENPM was significantly overexpressed in tumor tissue (Figure 5).
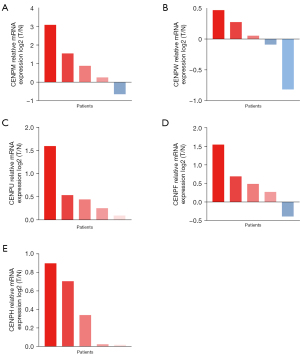
WGCNA construction and analysis of the correlation between the module and clinical characteristics
In TCGA LUAD data set (526 tumor tissues and 59 normal tissues), the median absolute value deviation (MAD) of hub genes was calculated, and the first 5,000 genes in terms of MAD value were selected to construct the WGCNA network. On the premise of maintaining appropriate network connectivity, the weighting factor β value was determined as 4 (Figure 6A,6B), and a total of 16 gene modules were established (Figure 6C). We then selected the modules related to clinical characteristics (age, gender, smoking, and stage), CENPF, CENPU, CENPM, CENPH, and CENPW. The turquoise module was positively correlated with CENPF (coefficient: 0.84; P=2e−137), CENPU (coefficient: 0.83; P=2e−133), CENPM (coefficient: 0.76; P=2e−99), CENPH (coefficient: 0.8; P=6e−115), and CENPW (coefficient: 0.84; P=6e−136) expressions. The brown module was negatively correlated with CENPF (coefficient: –0.57; P=6e−45), CENPU (coefficient: –0.62; P=4e−56), CENPM (coefficient: –0.58; P=7e−48), CENPH (coefficient: –0.56; P=9e−44), and CENPW (coefficient: –0.5; P=2e−34) expressions (Figure 6D). Genes in 2 modules may be regulated by the CENP family members. These results suggest that genes in the turquoise and brown modules may be regulated by CENPF, CENPU, CENPM, CENPH, and CENPW, and play an important role in the prognosis of patients with LUAD.
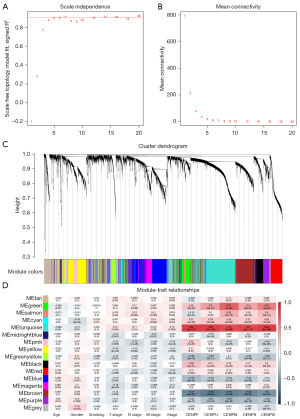
Functional enrichment analysis and identification of hub genes
To further explore the hub gene in the turquoise module, we selected the tumor tissue and normal tissue in TCGA LUAD data set and analyzed the differences between the 983 genes in the turquoise module. Finally, 162 downregulated genes and 279 upregulated genes were screened (Figure 7A). Next, we performed Gene Ontology (GO) enrichment analysis on 441 differential genes. Among them, the molecular function term suggested cofactor binding as the main function (gene ratio >0.04; P<0.05) (Figure 7B). In terms of biological process, it was enriched in mitotic sister chromosomal aggregation, DNA replication, sister chromosomal aggregation, nuclear chromosome aggregation, mitotic nuclear division, nuclear division, chromosome aggregation, and organelle fission (gene ratio >0.04; P<1e−13) (Figure 7C). The cell component term indicated that it was located in the kinetochore, chromosome, central region, microtubule, spindle, condensed chromosome, and chromosomal region (gene ratio >0.04; P<1e−07) (Figure 7D).
Discussion
An abundance of evidence indicates that CENPs not only participate in cell viability and mitosis but also are related to the progression and prognosis of several tumors (13,19,24,26). However, a comprehensive analysis of CENP expression and clinical significance in LUAD has not been conducted. In this study, we analyzed the transcriptional levels and their correlation with clinicopathological parameters, genetic alteration and coexpression pattern, potential function, and prognostic value of different CENP family members in LUAD. Fifteen CENP family proteins are differentially expressed in LUAD tissue. We used the LASSO algorithm to construct a risk model with 5 CENPs: CENPF, CENPU, CENPM, CENPH, and CENPW. The differential expression of these 5 CENP proteins was validated by qRT-PCR on the mRNA level, and WGCNA was performed to screen the genes related with these 5 CENP proteins. The molecular function term suggests cofactor binding as the main function. Our results can facilitate a more accurate individualized prediction for patients with LUAD and provide important guidance for the prognosis of the disease.
Various studies have indicated that CENPF is involved in the progression and metastasis of many cancers. For instance, the COUP-TFII-FOXM1-CENPF axis regulated by microRNA (miR)-101 and miR-27a contributes to the metastasis of prostate cancer (27). The HnRNPR-CCNB1/CENPF axis leads to the tumor proliferation and metastasis of gastric cancer (28). However, the role of CENPF in lung cancer remains unclear. Transcriptome analysis research suggests that CENPF acts as an oncogene in lung cancer (29). In this study, higher CENPF mRNA expression was observed in patients with LUAD, and the clinical characteristics were related. A short OS and progression-free survival (PFS) were also observed in patients with high CENPF expression.
Like CENPF, studies on CENPH in lung cancer are scarce. A study linking clinicopathologic characteristics with the CENPH expression pattern suggests that CENPH may be a prognostic biomarker for early NSCLC (24). Similar results were found in our study, and the potential function and mechanism of CENPH in LUAD were predicted for further research.
CENPM has attracted considerable attention due to its function in tumor progression (30,31). The upregulation of CENPM facilitates tumor metastasis in pancreatic cancer and promotes hepatocarcinogenesis. The results of our investigation demonstrate that CENPM was upregulated in LUAD tissue and related to tumor stage and nodal metastasis status. It may also be a predictive biomarker, as shorter OS and PFS were observed in patients with high CENPM expression.
The CENP-O-P-Q-U-R complex is part of the CCAN and participates in chromosome congression and oscillations throughout the cell cycle (14,32). A few studies have linked these proteins with tumors. One study indicated that CENPU was essential for papilloma development in a skin carcinogenesis model (33). In the current study, we found that CENPU was significantly upregulated in the tumor tissues of patients with LUAD. Individual tumor stage and nodal metastasis status were also related. Interestingly, the CENP-O/Q/U perform biological functions as a complex, since they compose the CENP-O-P-Q-U-R complex, which is a part of the CCAN. It thus seems that they share a similar biological process and may have a similar effect on tumor development.
CENPW is involved in the formation of the CENP-T-W-S-X complex, which directly binds to DNA and plays a crucial role in cell division during mitosis (14,34). We suggest that CENPW may be a potential biomarker of LUAD, as we found its expression level was increased in tumor tissues and related to clinical characteristics, and patients with high CENPW expression have a poor prognosis.
Some limitations to this study should be noted. First, a validation data set was lacking, so additional clinical data are needed to validate the model. Second, the interaction mechanism between the CENP proteins and downstream molecules needs to be further explored. Finally, future research should use a greater number of clinical samples to verify our findings.
Conclusions
The results of our study indicated that the mRNA expression levels of 13 CENP family members were upregulated in lung cancer tissues compared to those in normal tissues. CENPW, CENPM, CENPU, CENPF, and CENPH were significantly positively associated with prognosis in patients with LUAD. Furthermore, a poor OS prognosis was observed in patients with high mRNA expression of CENPs. According to the coexpression network of the 5 CENPs, 441 hub genes were screened, with their main function being cell mitosis. Future research will focus on the detailed mechanisms of the CENP family.
Acknowledgments
The research results are based on the data generated by TCGA Research Network and the Genotype-Tissue Expression (GTEx) database. We would like to thank Jimmy Zeng and his team for their selfless help in contributing to the bioinformatics analysis methods and thank Qi Tan for supporting us in our experiments.
Funding: This work was supported by the National Natural Science Foundation of China (No. 82072593) and Department of Science and Technology of Hubei Province (No. 2020BCB027).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2166/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2166/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2166/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2166/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Research Ethics Committee, Tongji Medical College, Huazhong University of Science and Technology (No. 20180403). Informed consent was waived due to the retrospective nature of this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Kim JW, Marquez CP, Kostyrko K, et al. Antitumor activity of an engineered decoy receptor targeting CLCF1-CNTFR signaling in lung adenocarcinoma. Nat Med 2019;25:1783-95. [Crossref] [PubMed]
- Chen H, Carrot-Zhang J, Zhao Y, et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat Commun 2019;10:5472. [Crossref] [PubMed]
- Nigro A, Ricciardi L, Salvato I, et al. Enhanced Expression of CD47 Is Associated With Off-Target Resistance to Tyrosine Kinase Inhibitor Gefitinib in NSCLC. Front Immunol 2020;10:3135. [Crossref] [PubMed]
- Bria E, Milella M, Cuppone F, et al. Outcome of advanced NSCLC patients harboring sensitizing EGFR mutations randomized to EGFR tyrosine kinase inhibitors or chemotherapy as first-line treatment: a meta-analysis. Ann Oncol 2011;22:2277-85. [Crossref] [PubMed]
- Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413-21. [Crossref] [PubMed]
- Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta 2008;1786:32-40. [PubMed]
- Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018;33:676-689.e3. [Crossref] [PubMed]
- Shrestha RL, Ahn GS, Staples MI, et al. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 2017;8:46781-800. [Crossref] [PubMed]
- Putkey FR, Cramer T, Morphew MK, et al. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell 2002;3:351-65. [Crossref] [PubMed]
- O'Brien SL, Fagan A, Fox EJ, et al. CENP-F expression is associated with poor prognosis and chromosomal instability in patients with primary breast cancer. Int J Cancer 2007;120:1434-43. [Crossref] [PubMed]
- Sharma AB, Dimitrov S, Hamiche A, et al. Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks. Nucleic Acids Res 2019;47:1051-69. [Crossref] [PubMed]
- Hinshaw SM, Harrison SC. Kinetochore Function from the Bottom Up. Trends Cell Biol 2018;28:22-33. [Crossref] [PubMed]
- McKinley KL, Cheeseman IM. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 2016;17:16-29. [Crossref] [PubMed]
- Hara M, Fukagawa T. Critical Foundation of the Kinetochore: The Constitutive Centromere-Associated Network (CCAN). Prog Mol Subcell Biol 2017;56:29-57. [Crossref] [PubMed]
- El-Arabey AA, Salama SA, Abd-Allah AR. CENP-E as a target for cancer therapy: Where are we now? Life Sci 2018;208:192-200. [Crossref] [PubMed]
- Shan L, Zhao M, Lu Y, et al. CENPE promotes lung adenocarcinoma proliferation and is directly regulated by FOXM1. Int J Oncol 2019;55:257-66. [PubMed]
- Srivastava S, Zasadzińska E, Foltz DR. Posttranslational mechanisms controlling centromere function and assembly. Curr Opin Cell Biol 2018;52:126-35. [Crossref] [PubMed]
- Smurova K, De Wulf P. Centromere and Pericentromere Transcription: Roles and Regulation … in Sickness and in Health. Front Genet 2018;9:674. [Crossref] [PubMed]
- Lu G, Shan T, He S, et al. Overexpression of CENP-H as a novel prognostic biomarker for human hepatocellular carcinoma progression and patient survival. Oncol Rep 2013;30:2238-44. [Crossref] [PubMed]
- He WL, Li YH, Yang DJ, et al. Combined evaluation of centromere protein H and Ki-67 as prognostic biomarker for patients with gastric carcinoma. Eur J Surg Oncol 2013;39:141-9. [Crossref] [PubMed]
- He P, Hu P, Yang C, et al. Reduced expression of CENP-E contributes to the development of hepatocellular carcinoma and is associated with adverse clinical features. Biomed Pharmacother 2020;123:109795. [Crossref] [PubMed]
- Wu Q, Chen YF, Fu J, et al. Short hairpin RNA-mediated down-regulation of CENP-A attenuates the aggressive phenotype of lung adenocarcinoma cells. Cell Oncol (Dordr) 2014;37:399-407. [Crossref] [PubMed]
- Liao WT, Wang X, Xu LH, et al. Centromere protein H is a novel prognostic marker for human nonsmall cell lung cancer progression and overall patient survival. Cancer 2009;115:1507-17. [Crossref] [PubMed]
- GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group. Genetic effects on gene expression across human tissues. Nature 2017;550:204-13.
- Sun X, Clermont PL, Jiao W, et al. Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int J Cancer 2016;139:899-907. [Crossref] [PubMed]
- Lin SC, Kao CY, Lee HJ, et al. Dysregulation of miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of prostate cancer. Nat Commun 2016;7:11418. [Crossref] [PubMed]
- Chen EB, Qin X, Peng K, et al. HnRNPR-CCNB1/CENPF axis contributes to gastric cancer proliferation and metastasis. Aging (Albany NY) 2019;11:7473-91. [Crossref] [PubMed]
- Meng F, Zhang L, Ren Y, et al. Transcriptome analysis reveals key signature genes involved in the oncogenesis of lung cancer. Cancer Biomark 2020;29:475-82. [Crossref] [PubMed]
- Xiao Y, Najeeb RM, Ma D, et al. Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms. J Exp Clin Cancer Res 2019;38:458. [Crossref] [PubMed]
- Zheng C, Zhang T, Li D, et al. Upregulation of CENPM facilitates tumor metastasis via the mTOR/p70S6K signaling pathway in pancreatic cancer. Oncol Rep 2020;44:1003-12. [Crossref] [PubMed]
- Kagawa N, Hori T, Hoki Y, et al. The CENP-O complex requirement varies among different cell types. Chromosome Res 2014;22:293-303. [Crossref] [PubMed]
- Saito M, Kagawa N, Okumura K, et al. CENP-50 is required for papilloma development in the two-stage skin carcinogenesis model. Cancer Sci 2020;111:2850-60. [Crossref] [PubMed]
- Chun Y, Lee M, Park B, et al. CSN5/JAB1 interacts with the centromeric components CENP-T and CENP-W and regulates their proteasome-mediated degradation. J Biol Chem 2013;288:27208-19. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


