
Assessment of the prognostic and clinicopathological significance of HOXA-AS2 in human cancers: a systematic review and meta-analysis
Highlight box
Key findings
• The high expression of HOXA cluster antisense RNA 2 (HOXA-AS2) is related to poor prognosis and unfavorable clinicopathological features in those with cancer, which suggests that HOXA-AS2 may serve as a promising prognostic marker for these patients.
What is known and what is new?
• HOXA-AS2 expression plays a role in cancer development and is associated with the prognosis of cancer patients.
• HOXA-AS2 could be used as an efficient prognostic marker for cancer patients.
What is the implication, and what should change now?
• More high-quality and large-sample studies need to be conducted to confirm the significant role of HOXA-AS2 in pan-cancer patients.
Introduction
Cancer is a leading cause of human morbidity and mortality. It represents a severe threat to human health and is a challenging public issue that needs to be resolved. The American Cancer Society estimated that there would be 1,918,030 new cancer cases and 609,360 cancer deaths in the United States in 2022 (1). Substantial progress has been made in the diagnosis and treatment of malignant tumors; however, the poor prognosis of cancer patients is still a major clinical problem (2). Thus, the search for new therapeutic targets and prognostic biomarkers is of significant clinical value for cancer patients.
With the rise of next-generation sequencing technologies, long noncoding RNA (lncRNA), which lacks specific open reading frames and has no protein-coding ability, has been found to be a key molecule in various diseases (3-5). It has been widely reported that lncRNAs are involved in many physiological and pathological processes, and play an important role in transcriptional regulation, cell scaffold assembly, protein localization, and chromatin modification (6-8). A growing number of studies have demonstrated that lncRNAs can act as oncogenes or tumor suppressor factors, and their abnormal expression is closely related to the prognosis and poor clinicopathological characteristics of patients with cancer, which suggests that lncRNAs have the potential to serve as new biomarkers for cancer prognosis (9-13).
HOXA cluster antisense RNA 2 (HOXA-AS2), an antisense lncRNA with a length of 1,048 bp, is located on the HOXA gene cluster. Numerous studies have shown that HOXA-AS2 is abnormally expressed in a variety of cancers, including gastric cancer (14), hepatocellular carcinoma (15), colorectal cancer (9), bladder cancer (16), and non-small cell lung cancer (NSCLC) (17), and is closely correlated with poor prognosis and poor clinical features in patients with cancer (18,19). It also plays a significant role in the development and progression of cancer by regulating the proliferation, migration, invasion, differentiation, and other biological behaviors of cancer cells (20,21). Due to inconsistencies in the findings of published studies, we performed a meta-analysis to evaluate the difference in the overall survival (OS), disease-free survival, and clinicopathological features among patients who were histologically diagnosed as having cancer with high or low HOXA-AS2 expression to clarify the prognostic and clinicopathological significance of HOXA-AS2 in a variety of human cancers. We present the following article in accordance with the PRISMA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1998/rc).
Methods
Search strategy
Databases, including PubMed, Web of Science, Embase, Wanfang Data, and China National Knowledge Infrastructure, were searched from January 2015 to September 2022 to retrieve studies examining the association between HOXA-AS2 expression and patient prognosis. The retrieval keywords were as follows: “(HOXA-AS2 or HOXA Cluster Antisense RNA 2) and (cancer or tumor or malignancy)”.
Literature screening
The literatures were screened according to the established inclusion and exclusion criteria. To be eligible for inclusion in this meta-analysis, the studies had to meet the following inclusion criteria: (I) include patients with a histopathological diagnosis, (II) include an intervention in which the expression level of HOXA-AS2 in the tissues of the patients was detected, (III) divide patients into a high expression group and low expression group based on the median or mean value, (IV) include sufficient data to calculate the hazard ratios (HRs) and odds ratios (ORs) and their 95% confidence intervals (CIs) so that the relationship between the expression of HOXA-AS2 and the prognosis and clinicopathological characteristics of patients with malignant tumors could be evaluated, and (V) have a retrospective analysis study design. The exclusion criteria for literature were the following: (I) case reports, meta-analyses, animal studies, or editorial comments; (II) a lack of sufficient data available; and (III) published in a language other than English or Chinese.
Data extraction and analysis
The data of the included studies were extracted separately by 2 researchers (BL and ZL), and a third researcher (XJ) helped to resolve any disagreements. The following information was extracted from the articles: first author, gender, age, publication date, cancer type, sample size, detection method, follow-up time, clinicopathological characteristics, and prognosis information. If the results of both the univariate and multivariate analyses were provided in a study, the data of the latter were used. If the HRs and 95% CIs were not provided directly in the articles, Engauge Digitizer (v. 10.4) was used to extract them from the Kaplan-Meier survival curve. The quality of the included articles was assessed using the Newcastle-Ottawa Scale (NOS) with scores ranging from 0 to 9; articles with scores >6 were considered high quality.
The Cancer Genome Atlas (TCGA) data analysis
The information in TCGA database was analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) web tool, and the relationship between HOXA-AS2 and the clinical stage of the patients was assessed using the following criteria: |log2 fold change| ≥1 and a P value ≤0.01. The correlation between the expression of HOXA-AS2 and the OS of the 9,481 patients was also analyzed using the above criteria in the survival analysis.
Immunoassays
Using the CIBERSORT algorithm, the RNA-sequencing data of patients in TCGA database were analyzed using the R package “immunedeconv” (The R Foundation for Statistical Computing) The correlations between HOXA-AS2 expression and the expression of sialic acid binding immunoglobin (Ig)-like lectin 15 a (SIGLEC15), T cell immunoreceptor with Ig and immunoreceptor tyrosine-based inhibitory motif (ITIM) domain protein (TIGIT), CD274, hepatitis A virus cellular receptor 2 (HAVCR2), programmed cell death 1 (PDCD1), cytotoxic T-lymphocyte–associated protein 4 (CTLA4), lymphocyte activating 3 (LAG3), and programmed cell death 1 ligand 2 (PDCD1LG2), which are the genes associated with immune checkpoints, were analyzed in a variety of cancers.
Statistical analysis
The statistical analysis was conducted using Stata software (v. 12.0, StataCorp). The HRs and ORs with 95% CIs were used to evaluate the relationship between HOXA-AS2 expression and prognosis and the clinicopathological parameters of patients with cancer. The heterogeneity among the included studies was evaluated using the Q and I2 tests. If there was no heterogeneity (I2<50% and P>0.05), a fixed-effects model was used; if there was heterogeneity (I2≥50% and P≤0.05), a random-effects model was used. Subgroup and sensitivity analyses were conducted to explore the source of heterogeneity. In addition, the Begg test was used to assess potential publication bias. A P value <0.05 was considered statistically significant.
Results
Studies selection and characteristics
Details of the article screening process are provided in Figure 1. A total of 374 articles were retrieved from the relevant databases. After exclusion of duplicate articles, reviews, and other unrelated literature, the remaining 45 articles underwent full-text readings, and 17 eligible studies were ultimately included in the meta-analysis. All the included studies were published between 2015 and 2021, and had NOS scores ranging from 6 to 8. The sample sizes of the studies ranged from 27 to 128. In total, 11 types of cancer were analyzed, including oral squamous cell carcinoma (OSCC) (18), hepatocellular carcinoma (15,22,23), prostate cancer (21), NSCLC (17,24,25), bladder cancer (16), glioma (26), osteosarcoma (27,28), papillary thyroid cancer (19,29), colorectal cancer (9), breast cancer (30), and gastric cancer (14). The specific characteristics of the included articles are set out in Table 1.
Table 1
| Study | Year | Country | Tumor type | Sample size (n) | HOXA-AS2 expression | Outcome information | HR methods | Follow-up, months | Laboratory method | NOS score | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | |||||||||||
| Chen | 2021 | China | Oral squamous cell carcinoma | 46 | 23 | 23 | – | – | – | qRT-PCR | 7 | (18) |
| Lu | 2020 | China | Hepatocellular carcinoma | 116 | 58 | 58 | OS | Directly | ≤60 | qRT-PCR | 7 | (15) |
| Xiao | 2020 | China | Prostate cancer | 68 | – | – | OS | Indirectly | ≤60 | qRT-PCR | 8 | (21) |
| Cui | 2019 | China | Non-small cell lung cancer | 40 | 20 | 20 | OS | Indirectly | >60 | qRT-PCR | 6 | (24) |
| Liu | 2019 | China | Non-small cell lung cancer | 52 | 27 | 25 | OS | Indirectly | ≤60 | qRT-PCR | 7 | (17) |
| Wang | 2019 | China | Bladder cancer | 80 | 40 | 40 | – | – | – | qRT-PCR | 8 | (16) |
| Wu | 2019 | China | Glioma | 50 | 25 | 25 | – | – | – | qRT-PCR | 7 | (26) |
| Wang | 2019 | China | Osteosarcoma | 27 | – | – | OS | Indirectly | ≤60 | qRT-PCR | 6 | (27) |
| Jiang | 2019 | China | Papillary thyroid cancer | 68 | 30 | 38 | OS | Indirectly | ≤60 | qRT-PCR | 6 | (19) |
| Wang | 2018 | China | Osteosarcoma | 66 | 33 | 33 | – | – | – | qRT-PCR | 7 | (28) |
| Xia | 2018 | China | Papillary thyroid cancer | 128 | 66 | 62 | – | – | – | qRT-PCR | 7 | (29) |
| Zhang | 2018 | China | Hepatocellular carcinoma | 58 | 38 | 20 | – | – | – | qRT-PCR | 7 | (22) |
| Ding | 2017 | China | Colorectal cancer | 69 | 35 | 34 | – | – | – | qRT-PCR | 8 | (9) |
| Li | 2017 | China | Non–small cell lung cancer | 103 | 52 | 51 | OS | Directly | >60 | qRT-PCR | 7 | (25) |
| Fang | 2017 | China | Breast cancer | 38 | – | – | OS | Indirectly | >60 | qRT-PCR | 8 | (30) |
| Wang | 2016 | China | Hepatocellular carcinoma | 112 | 56 | 56 | OS | Indirectly | >60 | qRT-PCR | 6 | (23) |
| Xie | 2015 | China | Gastric cancer | 55 | 28 | 27 | OS | Indirectly | ≤60 | qRT-PCR | 7 | (14) |
Indirectly: HR extracted from survival curve; directly: HR extracted from paper; –, not available. HR, hazard ratio; NOS, Newcastle-Ottawa scale; OS, overall survival; qRT-PCR, quantitative real time-polymerase chain reaction.
Relationship between the expression of HOXA-AS2 and the prognosis of patients with cancer
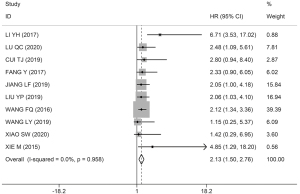
Among the 17 studies, 10 studies, comprising 669 patients, assessed the association between HOXA-AS2 expression levels and OS. As non-significant heterogeneity was found among the studies (I2=0.0%, P=0.958), the fixed-effects model was used. The results showed that the OS of patients in the HOXA-AS2 high-expression group was worse than that of patients in the HOXA-AS2 low-expression group (HR =2.13; 95% CI: 1.50–2.76; P≤0.001; Figure 2).
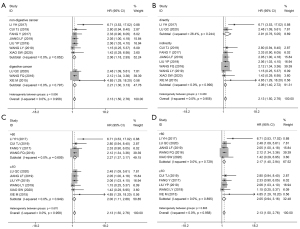
No heterogeneity was found among studies related to OS; however, we performed a subgroup analysis based on the cancer type, HR extraction method, follow-up month, and sample size to exclude potential biases. As Figure 3 shows, subgroup analysis demonstrated that the high expression of HOXA-AS2 could estimate unfavorable OS in the digestive system (I2=0.0%, P=0.852; HR =2.06, 95% CI: 1.18–2.93, P≤0.001; Figure 3A), nondigestive system (I2=0.0%, P=0.797; HR =2.21, 95% CI: 1.30–3.13, P≤0.001; Figure 3A), direct HR extraction (I2=26.4%, P=0.244; HR =2.91, 95% CI: 0.76–5.05, P≤0.001; Figure 3B), indirect HR extraction (I2=0.0%, P=0.990; HR =2.06, 95% CI: 1.40–2.72, P≤0.001; Figure 3B), sample size >60 (I2=0.0%, P=0.609; HR =2.27, 95% CI: 1.37–3.17, P≤0.001; Figure 3C), sample size ≤60 (I2=0.0%, P=0.949; HR =2.00, 95% CI: 1.11–2.88; P≤0.001; Figure 3C), follow-up month >60 (I2=0.0%, P=0.729; HR =2.17, 95% CI: 1.40–2.94, P≤0.001; Figure 3D), follow-up ≤60 months (I2=0.0%, P=0.895; HR =2.05, 95% CI: 0.94–3.16, P≤0.001; Figure 3D). These results revealed no significant bias in the study related to OS.
Relationship between the expression of HOXA-AS2 and the clinicopathological features
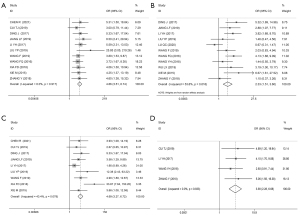
ORs and 95% CIs were used to evaluate the relationship between HOXA-AS2 expression and patients’ clinicopathological characteristics. As Figure 4 shows, the results revealed that elevated HOXA-AS2 expression was significantly correlated with high tumor stage (HTS) (OR =4.86, 95% CI: 3.51–6.74, P≤0.001; I2=0.0%, P=0.917; Figure 4A), large tumor size (LTS) (OR =2.33, 95% CI: 1.51–3.60; P≤0.001; I2=53.6%, P=0.018; Figure 4B), lymph node metastasis (LNM) (OR =4.69, 95% CI: 3.27–6.72, P≤0.001; I2=43.4%, P=0.078; Figure 4C), and distant metastasis (DM) (OR =3.90, 95% CI: 2.28–6.68, P≤0.001; I2=0.0%, P=0.833; Figure 4D). However, high HOXA-AS2 expression was not related to a poor histological grade (PHG) (OR =1.59, 95% CI: 0.92–2.74, P=0.094; I2=53.3%, P=0.058), age (OR =1.07, 95% CI: 0.84–1.37, P=0.564; I2=0.0%, P=0.551), or gender (HR =0.92, 95% CI: 0.70–1.21, P=0.548; I2=4.6%, P=0.401).
Sensitivity analysis and publication bias
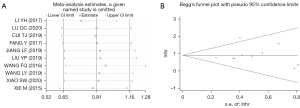
In the study of OS, the sensitivity analysis showed that the overall results were not significantly affected when the results of any single study were deleted (Figure 5A). The Begg test results also revealed no publication bias among the studies examining OS (Z =0.36; probability > |z| =0.721; Figure 5B).
TCGA database analysis
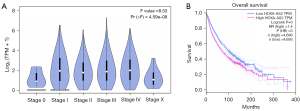
In this study, the GEPIA web tool was used to analyze TCGA data to further validate the above results. The results showed that the expression of HOXA-AS2 was significantly correlated with the clinical stage of patients (F =8.53; probability (> F) ≤0.001; Figure 6A). Importantly, the survival analysis indicated that patients with high HOXA-AS2 expression had shorter OS and a poorer prognosis than those with low HOXA-AS2 expression (HR =1.4; P≤0.001; n=9,389; Figure 6B).
Immunoassay
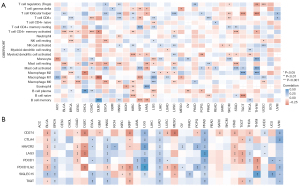
HOXA-AS2 was found to be significantly correlated with immune cell infiltration in a variety of cancers. For example, in bladder cancer, the expression of HOXA-AS2 was significantly and positively correlated with activated mast cells and memory-resting CD4+ T cells, and significantly negatively correlated with memory-activated CD4+ T cells and resting mast cells (Figure 7A). In uterine corpus endometrial carcinoma, the expression of HOXA-AS2 was significantly and positively correlated with activated natural killer cells and M1 macrophage infiltration, and significantly negatively correlated with regulatory T-cell and M0 macrophage infiltration. Additionally, the immune checkpoint analysis showed that in lung squamous cell carcinoma, the expression of HOXA-AS2 was significantly and positively correlated with CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, SIGLEC15, and TIGIT checkpoints, while in gastric cancer, the expression of HOXA-AS2 was significantly and negatively correlated with CD274, HAVCR2, LAG3, and PDCD1 checkpoints (Figure 7B). The above results may provide new directions for studying the immunoregulatory mechanisms and immunotherapeutic targets involved in HOXA-AS2 in cancer.
Discussion
Cancers are not only a great threat to human health, but also place a serious burden on the global economy (31). Despite many significant advances, the prognosis of patients with cancer is still poor, which is mainly due to the lack of effective diagnostic markers and therapeutic targets (32). Previous studies have confirmed that lncRNAs can affect the occurrence and progression of tumors by regulating the proliferation, migration, invasion, differentiation, and apoptosis of cancer cells, and lncRNAs can be considered promising prognostic markers of cancer (33,34).
There is mounting evidence that lncRNA HOXA-AS2 is upregulated in a variety of cancers and is closely related to the poor prognosis and clinicopathological characteristics of patients with cancer (16,23,28). Thus, this meta-analysis sought to further evaluate the potential of HOXA-AS2 as a prognostic marker for cancer patients. A total of 17 articles, comprising 1176 patients and 11 different types of cancer, were included in this meta-analysis. The results showed that the elevated expression of HOXA-AS2 was significantly associated with the poor OS of those with tumor, and the subgroup analysis and Begg test results further verified the reliability of these results. In addition, patients with high expressions of HOXA-AS2 were vulnerable to LNM, LTS, HTS, and DM. However, the high expression of HOXA-AS2 was not related to the age, gender, or the PHG of patients. Moreover, consistent with above results, the analysis of TCGA data showed that the elevated expression of HOXA-AS2 is associated with poor OS and clinical stage in those with tumor, which further validated the results of our meta-analysis.
Numerous researchers have shown that the competing endogenous RNA network plays an important role in the regulation of lncRNAs in cancer (28,29). Chen et al. found that increased HOXA-AS2 expression suppresses the proliferation OSCC cells by inhibiting miR-567 (18). Cui et al. showed that HOXA-AS2 targets miR-216a-5p and thus affects the proliferation, progression, migration, and epithelial–mesenchymal transition process of NSCLC cells (24). Liu et al. also revealed that the high expression of HOXA-AS2 is related to the poor prognosis of patients with NSCLC through the regulation of proliferation, apoptosis, and migration of cancer cells via the repression of miR-520a-3p (17). In breast cancer, the inhibition of HOXA-AS2 expression was shown to promote cell-cycle arrest and apoptosis by competitively binding to miR-520c-3p, thereby inhibiting the proliferation, migration, and invasion of cancer cells (30).
Immunotherapy, such as chimeric antigen receptor T-cell immunotherapy, cell therapy, and immune checkpoint inhibitor programmed cell death protein 1 (PD-1), has shown great potential in the treatment of patients with cancer and has been proven to have good efficacy (35). Thus, this study performed an immune correlation analysis of HOXA-AS2 in cancers, and the results revealed a significant correlation between the expression of HOXA-AS2 and infiltration of immune cells and immune checkpoints in cancers. Thus, the above regulatory mechanisms found in this study may provide a new direction for studying the role of HOXA-AS2 in cancers.
Our study had some limitations. First, the included patients were all Chinese; thus, our findings may not apply to other races. Second, some HRs and 95% CIs were indirectly extracted from the survive curve, which might have affected the validity of our findings. Finally, the sample size of our study was relatively small, and further studies need to be conducted to comprehensively evaluate the prognostic role of HOXA-AS2 in patients with cancer.
Conclusions
The high expression of HOXA-AS2 is associated with poor prognosis and unfavorable clinicopathological features in patients with cancer, which suggests that HOXA-AS2 may serve as a promising prognostic marker for these patients. More high-quality, large-sample studies need to be conducted to further validate our findings.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1998/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1998/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1998/coif). The authors have no conflicts of interest to declare except the funding.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Miller KD, Nogueira L, Devasia T, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin 2022;72:409-36. [Crossref] [PubMed]
- Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov 2021;20:629-51. [Crossref] [PubMed]
- Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem 2021;65:625-39. [Crossref] [PubMed]
- Oo JA, Brandes RP, Leisegang MS. Long non-coding RNAs: novel regulators of cellular physiology and function. Pflugers Arch 2022;474:191-204. [Crossref] [PubMed]
- Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016;539:452-5. [Crossref] [PubMed]
- Zhou B, Yang H, Yang C, et al. Translation of noncoding RNAs and cancer. Cancer Lett 2021;497:89-99. [Crossref] [PubMed]
- Dahariya S, Paddibhatla I, Kumar S, et al. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol Immunol 2019;112:82-92. [Crossref] [PubMed]
- Ding J, Xie M, Lian Y, et al. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis 2017;6:e288. [Crossref] [PubMed]
- Wang H, Liu Y, Tang A. Prognostic Values of Long Noncoding RNA linc00152 in Various Carcinomas: An Updated Systematic Review and Meta-Analysis. Oncologist 2020;25:e31-8. [Crossref] [PubMed]
- Tian S, Yu Y, Huang H, et al. Expression Level and Clinical Significance of NKILA in Human Cancers: A Systematic Review and Meta-Analysis. Biomed Res Int 2020;2020:4540312. [Crossref] [PubMed]
- Zhao S, Zhu H, Jiao R, et al. Prognostic and clinicopathological significance of SNHG6 in human cancers: a meta-analysis. BMC Cancer 2020;20:77. [Crossref] [PubMed]
- Yang Q, Wang J, Zhong P, et al. The clinical prognostic value of lncRNA FAM83H-AS1 in cancer patients: a meta-analysis. Cancer Cell Int 2020;20:72. [Crossref] [PubMed]
- Xie M, Sun M, Zhu YN, et al. Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget 2015;6:33587-601. [Crossref] [PubMed]
- Lu Q, Gao J, Tang S, et al. Integrated RNA Sequencing and Single-Cell Mass Cytometry Reveal a Novel Role of LncRNA HOXA-AS2 in Tumorigenesis and Stemness of Hepatocellular Carcinoma. Onco Targets Ther 2020;13:10901-16. [Crossref] [PubMed]
- Wang F, Wu D, Chen J, et al. Long non-coding RNA HOXA-AS2 promotes the migration, invasion and stemness of bladder cancer via regulating miR-125b/Smad2 axis. Exp Cell Res 2019;375:1-10. [Crossref] [PubMed]
- Liu Y, Lin X, Zhou S, et al. Long noncoding RNA HOXA-AS2 promotes non-small cell lung cancer progression by regulating miR-520a-3p. Biosci Rep 2019;39:BSR20190283. [Crossref] [PubMed]
- Chen R, Wang X, Zhou S, et al. LncRNA HOXA-AS2 Promotes Tumor Progression by Suppressing miR-567 Expression in Oral Squamous Cell Carcinoma. Cancer Manag Res 2021;13:5443-55. [Crossref] [PubMed]
- Jiang L, Wu Z, Meng X, et al. LncRNA HOXA-AS2 Facilitates Tumorigenesis and Progression of Papillary Thyroid Cancer by Modulating the miR-15a-5p/HOXA3 Axis. Hum Gene Ther 2019;30:618-31. [Crossref] [PubMed]
- Wang J, Su Z, Lu S, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta 2018;485:229-33. [Crossref] [PubMed]
- Xiao S, Song B. LncRNA HOXA-AS2 promotes the progression of prostate cancer via targeting miR-509-3p/PBX3 axis. Biosci Rep 2020;40:BSR20193287. [Crossref] [PubMed]
- Zhang Y, Xu J, Zhang S, et al. HOXA-AS2 Promotes Proliferation and Induces Epithelial-Mesenchymal Transition via the miR-520c-3p/GPC3 Axis in Hepatocellular Carcinoma. Cell Physiol Biochem 2018;50:2124-38. [Crossref] [PubMed]
- Wang F, Yang H, Deng Z, et al. HOX Antisense lincRNA HOXA-AS2 Promotes Tumorigenesis of Hepatocellular Carcinoma. Cell Physiol Biochem 2016;40:287-96. [Crossref] [PubMed]
- Cui TJ, Lin GS, Dai YM, et al. LncRNA HOXA-AS2 regulates microRNA-216a-5p to promote malignant progression of non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2019;23:264-73. [PubMed]
- Li Y, Jiang H. Up-regulation of long non-coding RNA HOXA-AS2 in non-small cell lung cancer is associated with worse survival outcome. Int J Clin Exp Pathol 2017;10:9690-6. [PubMed]
- Wu L, Zhu X, Song Z, et al. Long Non-Coding RNA HOXA-AS2 Enhances The Malignant Biological Behaviors In Glioma By Epigenetically Regulating RND3 Expression. Onco Targets Ther 2019;12:9407-19. [Crossref] [PubMed]
- Wang L, Wang L, Zhang X. Knockdown of lncRNA HOXA-AS2 Inhibits Viability, Migration and Invasion of Osteosarcoma Cells by miR-124-3p/E2F3. Onco Targets Ther 2019;12:10851-61. [Crossref] [PubMed]
- Wang Y, Zhang R, Cheng G, et al. Long non-coding RNA HOXA-AS2 promotes migration and invasion by acting as a ceRNA of miR-520c-3p in osteosarcoma cells. Cell Cycle 2018;17:1637-48. [Crossref] [PubMed]
- Xia F, Chen Y, Jiang B, et al. Long Noncoding RNA HOXA-AS2 Promotes Papillary Thyroid Cancer Progression by Regulating miR-520c-3p/S100A4 Pathway. Cell Physiol Biochem 2018;50:1659-72. [Crossref] [PubMed]
- Fang Y, Wang J, Wu F, et al. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget 2017;8:46090-103. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Davies A, Lum C, Raju R, et al. Anti-cancer therapy made easier: a 25-year update. Intern Med J 2021;51:473-80. [Crossref] [PubMed]
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21:22-36. [Crossref] [PubMed]
- Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer 2020;19:167. [Crossref] [PubMed]
- McGowan E, Lin Q, Ma G, et al. PD-1 disrupted CAR-T cells in the treatment of solid tumors: Promises and challenges. Biomed Pharmacother 2020;121:109625. [Crossref] [PubMed]


