
Identification of key genes in HER2-positive breast cancer with brain metastasis via bioinformatics methods
Highlight box
Key findings
• CD44, COL1A2, MMP14, POSTN, and SOX9 were HER2-positive BCBM associated genes, and were found to be related to the survival outcomes of HER2-positive patients.
What is known and what is new?
• HER2-positive BC is one of the most common BC with BM. Some BMs and paired primary BC shows different genomic characteristics, and these differences shown on genes can be applied to modulate the management of BCBM.
• HER2-positive BCBM associated genes were screened and their clinical significance in BC was verified.
What is the implication, and what should change now?
• CD44, COL1A2, MMP14, POSTN, and SOX9 are HER2-positive BCBM associated gene, and they are potential therapeutic targets. Further investigations are necessary to unravel the mechanisms.
Introduction
According to the International Agency for Research on Cancer, breast cancer (BC) is the most diagnosed malignancy worldwide (1). Brain metastasis (BM) is commonly observed in advanced BC patients with around 30% to 50% of metastatic BC (MBC) patients developing BM (2,3). The risk of BM as the first site of metastasis is low in stage I–II BC patients (4,5), but significantly increases in BC patients with stage III disease (6,7). In BC, 50% of human epidermal growth factor receptor 2 (HER2)-positive BC patients, 25% to 46% of triple-negative BC (TNBC) patients, and 10–15% of estrogen receptor (ER)-positive HER2-negative BC patients develop BM during their lifetime (8-11).
Although BC with BM (BCBM) patients can benefit from local treatments such as stereotactic radiosurgery (SRS), surgery, and to a lesser extent whole-brain radiation therapy (WBRT), the median survival time of patients with BM is only 3 to 27 months (12-14). Advancements in systemic management developed since the last two decades, which have improved outcomes of MBC patients, and the current median survival time of HER2-positive MBC patients is 5 years (15,16). The poor efficacy of anti-cancer drugs against metastases in the central nervous system (CNS) is mainly attributed to the blood–brain barrier, which prevents significant concentration of high molecular-weight drugs entering the brain tissues (17). Progressive CNS disease accounts for the mortality of 50% of HER2-positive BCBM patients (18). Whole exome sequencing of 86 matched BMs, primary tumors, and normal tissues showed that clinically relevant genomic characteristics were significantly different in 53% of paired brain metastatic samples and primary BC sample (19). Therefore, understanding the precise molecular mechanisms underlying BMs in HER2-positive BC patients is critical in developing effective BM-specific treatment strategies.
In this study, the microarray data of the HER2-positive primary BC tissues and HER2-positive BM tissues of BC patients from the Gene Expression Omnibus (GEO) database were analyzed to identify candidate BM-related biomarker genes that are associated with the survival outcomes of HER2-positive BCBM patients. We present this article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2715/rc).
Methods
Microarray data
The GEO database (https://www.ncbi.nlm.nih.gov/geo) is a public functional genomics data repository of the National Center for Biotechnology Information (NCBI) (20). The gene expression data of 19 BM tissues of HER2-positive BC patients and 19 HER2-positive non-metastatic primary BC samples were downloaded from the GSE43837 dataset (21). The probes in the Affymetrix Human X3P Array were converted into the corresponding gene symbol based on the annotation information in the platform.
Identification of differentially expressed genes (DEGs)
The GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r) web tool was used to identify the DEGs between BM tissues of HER2-positive BC and nonmetastatic primary HER2-positive BC samples. Benjamini & Hochberg (false discovery rate) was used to compare differences of genes expression in two groups. |log(fold change) (logFC)| >1 and P value <0.01 were considered as threshold parameters.
Gene ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs
DAVID (https://david.ncifcrf.gov/) (version 6.8) is a database for the functional annotation of large lists of genes (22,23). GO analysis is used to identify the molecular functions (MFs), biological processes (BPs), and cellular components (CCs) associated with the DEGs (24). KEGG database is used for systematic analysis of high-level gene functions and pathways (25). DAVID (version 6.8) from the Bioinformatics website (https://www.bioinformatics.com.cn/) was applied for visualizing the results of the GO and KEGG pathway enrichment analysis. P<0.05 was considered statistically significant.
Protein-protein interaction (PPI) network construction and analysis
STRING (https://string-db.org/) database is used to construct PPI networks (26). In the present study, STRING (version 11.5) database was used to construct a PPI network of DEGs; combined score >0.4 was considered statistically significant. Cytoscape (version 3.7.1) database is used for visualizing the molecular interaction networks (27). MCODE is an application in Cytoscape for identifying densely connected regions in a PPI network based on topology (28). MCODE uses vertex weighting, which is based on the clustering coefficient (Ci). It calculates the neighbor cliquishness Ci = 2n/ki(ki − 1) of a vertex, where n is the number of edges in the neighbor and ki is the vertex size of the neighbor of the vertex. MCODE selection were as follows: MCODE scores >5, degree cut-off =2, node score cut-off =0.2, maximum depth =100, and k-score =2.
Clinical significance analysis
UALCAN database (https://ualcan.path.uab.edu/) was used to perform in-depth analysis of the candidate DEGs by comparing the gene expression levels between 114 normal breast tissues and 1,097 primary BC tissues from breast invasive carcinoma dataset of The Cancer Genome Atlas (TCGA) database (29,30). Furthermore, the gene expression levels between 556 luminal subtype, 37 HER2-positive, 116 TNBC and 114 normal breast tissues from breast invasive carcinoma dataset of TCGA database were compared. The expression levels of candidate genes between 83 HER2-negative and 9 HER2-positive MBC tissues from MBC dataset of TCGA database were also analyzed.
Kaplan-Meier plotter tool (https://kmplot.com/analysis/) was used to assess the survival outcomes of 1,273 HER2-positive BC patients based on the expression levels of the candidate genes. In this study, the association between expression of candidate genes and outcomes of HER2-positive BC patients with or without BM was assessed.
Statistical analysis
Wilcoxon rank sum test was used to compare gene expression differences between groups. Kaplan-Meier survival analysis was used to compare survival differences in two groups. P<0.05 was considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Identification of 1,056 DEGs in BM samples of HER2-positive BC
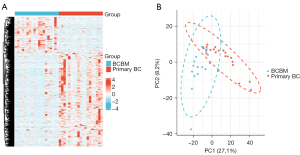
A total of 1,056 DEGs including 767 downregulated and 289 upregulated genes were identified by analyzing the transcriptome data of 19 BM samples of HER2-positive BC and 19 nonmetastatic primary BC samples from the GSE43837 dataset. The heatmap of these DEGs is shown in Figure 1A. Principal component analysis (PCA) is used for efficient dimensionality reduction and exploratory visualization of the data from the GSE43837 dataset. The BCBM group and primary BC group displayed different gene patterns (Figure 1B).
Functional enrichment analyses of DEGs
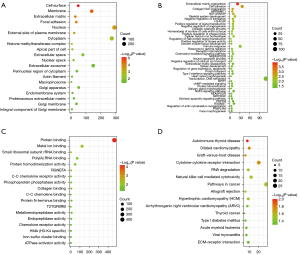
Functional enrichment analysis was performed using DAVID to identify biological mechanisms associated with the DEGs. GO analysis results showed that the DEGs were enriched in CC such as cell surface, membrane, extracellular matrix (ECM), focal adhesion, nucleus, and external side of plasma membrane (Figure 2A). Furthermore, DEGs were enriched in BP such as ECM organization, cell adhesion, collagen fibril organization, antigen processing and presentation of peptide antigen via major histocompatibility complex (MHC) class I, negative regulation of transforming growth factor beta receptor signaling pathway, and skeletal system development (Figure 2B). DEGs were also enriched in MF such as protein binding, metal ion binding, and small ribosomal subunit ribosomal RNA (rRNA) binding (Figure 2C). KEGG pathway analysis showed that the DEGs were enriched in pathways associated with autoimmune thyroid disease, dilated cardiomyopathy, graft-versus-host disease (Figure 2D).
After removing above one outlier in BCBM group and three outliers in primary BC group discovered by PCA analysis, GO analysis and KEGG pathway analysis were performed again. GO analysis results showed that the DEGs were enriched in CC such as ECM, membrane, cytosol, cell surface, and extracellular exosome (Figure S1A). Furthermore, DEGs were enriched in BP such as ECM organization, skeletal system development, positive regulation of protein catabolic process, embryonic limb morphogenesis, and osteoclast differentiation (Figure S1B). DEGs were also enriched in MF such as protein binding, platelet-derived growth factor binding, protein kinase A binding, protease binding, and integrin binding (Figure S1C). KEGG pathway analysis showed that the DEGs were enriched in pathway of neurodegeneration-multiple diseases, amoebiasis, rheumatoid arthritis, dilated cardiomyopathy, and advanced glycation end product (AGE)-receptor for AGE (RAGE) signaling pathway in diabetic complications (Figure S1D).
PPI network and module construction to identify hub genes
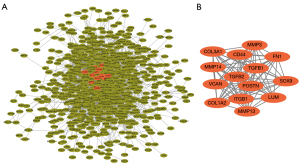
PPI network analysis of the DEGs was performed using the STRING 11.5 database (Figure 3A). Fourteen DEGS in the most significant module as visualized using the MCODE plugin of Cytoscape 3.7.1 (Figure 3B). Among these 14 DEGs, SOX9 was upregulated and the remaining 13 DEGs (FN1, COL1A2, COL3A1, MMP3, MMP13, MMP14, POSTN, VCAN, TGFB1, LUM, CD44, ITGB1, and ITGB2) were downregulated.
After removing above outliers, PPI network analysis was also performed (Figure S2A). Thirteen DEGs were identified by the MCODE plugin of Cytoscape 3.7.1 (Figure S2B). Among these 13 DEGs, SOX9 and ACTB were upregulated and the remaining 11 DEGs (COL1A1, COL1A2, COL3A1, MMP3, MMP13, MMP14, POSTN, VCAN, TGFB2, TGFB1, LUM, and ITGB1) were downregulated.
Clinical significance analysis
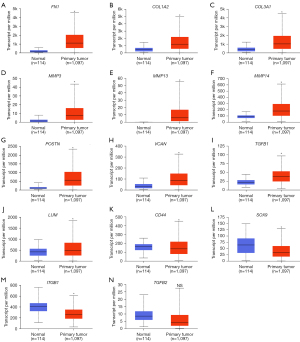
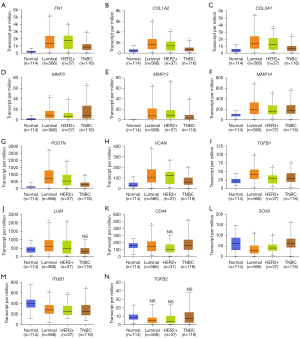
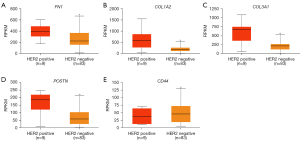
UALCAN was used to compare the expression levels of the 14 DEGs in clinical BC samples. Firstly, the expression levels of the 14 DEGs between 114 normal breast tissues and 1,097 primary BC tissues from breast invasive carcinoma dataset of TCGA database was compared. The results showed upregulation of FN1, COL1A2, COL3A1, MMP3, MMP13, MMP14, POSTN, VCAN, TGFB1, and LUM, and downregulation of CD44, SOX9, and ITGB1 in the primary BC samples compared to the normal breast tissue samples (Figure 4). The differences in the expression levels of the 14 DEGs between normal breast tissues and different BC subtypes were then analyzed. The results showed upregulation of FN1, COL1A2, COL3A1, MMP3, MMP13, MMP14, POSTN, VCAN, and TGFB1 in all the subtypes of BC, and upregulation of LUM in the luminal and HER2-positive BC tissue samples (Figure 5). Furthermore, SOX9 and ITGB1 were downregulated in all subtypes of BC, whereas CD44 was downregulated in the luminal and TNBC tissue samples (Figure 5). Finally, the differences in the gene expression levels of the 14 DEGs between 83 HER2-negative MBC and 9 HER2-positive MBC tissue samples was compared. The results showed higher expression of FN1, COL1A2, COL3A1, and POSTN and reduced expression of CD44 in the HER2-positive MBC tissue samples compared to the HER2-negative MBC tissue samples (Figure 6). The other 9 candidate genes had no differences between HER2-positive MBC and HER2-negative MBC tissue samples in current study (Figure S3).
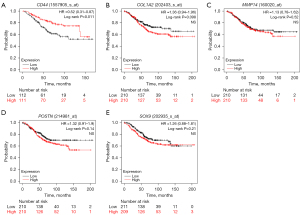
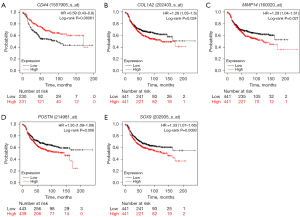
Kaplan-Meier survival curve analysis was performed to determine the differences in the overall survival (OS) and relapse-free survival (RFS) of the 1,273 HER2-positive BC patients based on the expression levels of the 14 candidate genes. HER2-positive patients with lower expression of CD44 showed significantly lower OS (P=0.011, Figure 7A) than those with high CD44 expression. Furthermore, OS rates were significantly lower for the HER2-positive BC patients with higher expression levels of COL1A2 (P=0.098, Figure 7B), MMP14 (P=0.52, Figure 7C), POSTN (P=0.14, Figure 7D), and SOX9 (P=0.21, Figure 7E) compared to those with lower expression of the corresponding genes. HER2-positive patients with lower expression of CD44 also showed significantly lower RFS (P=0.00061, Figure 8A) than those with high CD44 expression. Furthermore, RFS rates were also significantly lower for the HER2-positive BC patients with higher expression levels of COL1A2 (P=0.024, Figure 8B), MMP14 (P=0.021, Figure 8C), POSTN (P=0.006, Figure 8D), and SOX9 (P=0.0093, Figure 8E) compared to those with lower expression of the corresponding genes. The survival outcomes of the HER2-positive patients did not show any statistically significant differences based on the differential expression levels of the remaining 9 candidate genes.
Discussion
Nearly 30–50% of patients with MBC develop BM (2,3). Furthermore, the risk of BM is higher in patients with HER2-positive/hormone receptor (HR)-negative and TNBC (3). The mortality rate is 50% for HER2-positive BC patients with BM progression (18). In previous studies, researchers fully elucidated the gene expression patterns of BCBM (31,32). However, their results are not independent of HER2 status which will affect the gene expression profiles and management strategies. Although Kuroiwa et al. (33) has explored the expression signature of BM in HER2-positive BC, the screening processes are based on cell line and mouse xenograft which are different from the microenvironment of human BCBM. One study based on bioinformatic analysis has identified some hub genes in BM of HER2 positive BC (34). However, the genes they screened are not verified by data from other databases, to some extent leading to unreliability of their clinical application.
In present study, microarray analysis of a GEO BC dataset identified 1,056 DEGs, including 767 downregulated genes and 289 upregulated genes between BM tissues of HER2-positive BC and primary BC tissues. Functional enrichment analysis of the DEGs showed enrichment of pathways related to ECM organization, cell adhesion, and collagen fibril organization. Furthermore, 14 hub genes were identified as candidate genes regulating BM by the hierarchical clustering and PPI network analysis of the DEGs. Among these, SOX9 was upregulated and the remaining 13 DEGs were downregulated. PCA plot was also performed, and the BCBM group and primary BC group displayed different gene patterns. After removed one outliner in BCBM group and three outliers in primary BC group, GO analysis and KEGG pathway analysis was performed, and PPI network was constructed again. As functional enrichment is largely consistent, and most of the hub genes are overlapped, outlier samples were considered to be caused by tumor heterogeneity.
CD44 is an integral membrane protein that plays a pivotal role in cellular signaling and cell-cell communication, and also acts as a link between the ECM components and the intracellular cytoskeletal proteins (35,36). CD44 is a commonly used cancer stem cell (CSC) marker. CD44 overexpression is associated with cancer cell proliferation, metastasis, invasion, migration and stemness, and tumor resistance to chemotherapy and/or radiotherapy in several cancers (37). The relationship between CD44 expression levels and the clinicopathological features and survival outcomes of BC patients is not clear. In the present study, CD44 expression was significantly lower in the HER2-positive BCBM patients compared to the primary HER2-positive BC patients. Furthermore, CD44 was downregulated in BC, especially in the luminal BC and TNBC. Moreover, CD44 expression was significantly lower in the HER2-positive MBC compared to the HER2-negative MBC. OS and RFS rates were significantly worse for HER2-positive BC patients with low CD44 expression compared to those with higher CD44 expression.
The metastatic cascade involves local invasion through the basement membrane and the surrounding ECM, intravasation into the vessel or the lymphatic vessels, and subsequent dissemination to the distant sites (38). Epithelial-mesenchymal transition (EMT) and ECM remodeling are required for the initiation of cancer metastasis (39). Collagen is a major component of the ECM that is also involved in the development of human placenta (40). Matrix metalloproteinases (MMPs) are a family of calcium and zinc dependent proteases that play a key role in degrading the ECM proteins (41). MMP13 was first identified in BC as a key player in the activation cascade of the extracellular MMPs and ECM degradation (42). MMP13 promotes the initiation, growth, migration, and invasion of BC cells and is associated with aggressive BC phenotypes and poorer survival outcomes (43,44). In the animal models, MMP13 promoted lung metastasis and played a key role in the differentiation and activation of osteoclasts during BM (45,46). MMP3 is another MMP that is downregulated in BCBM compared to the primary BC tissues (47). However, MMP3 activity was higher in metastatic brain tissues compared to the normal brain tissues (48). The expression levels of MMP3 and MMP14 in brain-derived clones of MDA-MB-231 did not show significant differences compared to the normal brain tissues (49). MMP14 (also known as membrane type 1-MMP) is an activator of MMP2 and promotes cancer cell invasion, metastasis and angiogenesis by degrading ECM and cell adhesion proteins (50,51). MMP14 is downregulated in BCBM and associated with metastasis and poorer survival outcomes (52). In the present study, MMP3, MMP13, and MMP14 were upregulated in all the subtypes of BC, and their expression levels were independent of the HER2 status in MBC. HER2-positive BC patients with high MMP14 expression were associated with poorer RFS.
COL1A2 and COL3A1 encode the α2 chain of type I collagen and the α1 chain of types I and III collagens, respectively; moreover, type I and III collagens are important components of the ECM (53,54). Type I and III collagens participate in tumor invasion and progression (54-56). Furthermore, aberrant expression of COL1A2 is associated with survival outcomes in several cancers (57-61). The upregulation of COL1A2 and COL3A1 is associated with poor survival outcomes in patients with ER-positive BC (62,63). Furthermore, the expression levels of COL1A2 and COL3A1 are upregulated in BC after radiotherapy (64). A previous study showed that low expression of COL3A1 correlated with metastasis in patients with BCBM and poor survival outcomes (52). In this current study, COL1A2 and COL3A1 were downregulated in HER2-positive BCBM patients. However, further investigation showed that COL1A2 and COL3A1 were upregulated in all the subtypes of BC. Among MBC patients, HER2-positive BC group showed higher expression levels of COL1A2 and COL3A1. Furthermore, HER2-positive BC patients with higher COL1A2 expression showed poor RFS and OS compared to those with lower COL1A2 expression.
ECM proteins and secretory factors mediate tumor-stromal interactions (65). POSTN, also known as OSF-2, is a secretory cell-adhesion glycoprotein in the ECM that plays a critical role in tumor cell proliferation, adhesion, migration, and EMT (66). POSTN is also implicated in cancer cell stemness and regulates tumor angiogenesis, lymph-angiogenesis, and distant metastases (67,68). POSTN is highly expressed in the tumor stromal cells such as the cancer-associated fibroblasts in BC, and its upregulation correlates with tumor malignancy and shorter survival rates of the IDC patients (68-70). A positive feedback loop between POSTN and TGF-β promotes and maintains the stemness of cancer cells, and is associated with increased invasion and worse survival outcomes (71-73). In the GEO-BC dataset, POSTN and TGFB2 were downregulated in the HER2-positive BM. However, in further studies, POSTN was upregulated in all the subtypes of BC.
SOX9 is a member of the SOX family of transcription factors and is associated with tumorigenesis and poor survival outcomes in solid tumors (74). In non-small cell lung cancer (NSCLC), tumor-associated macrophages (TAMs) secrete TGF-β, which induces SOX9 expression via the c-Jun/SMAD3 pathway and subsequently promotes tumor progression and metastasis (75). SOX9 promotes BC growth, proliferation, migration, invasion, and metastasis by directly regulating genes involved in cellular apoptosis and EMT (76-78). Sox9 upregulation is associated with the CD44+/CD24−/low phenotype and poor prognosis in BC (79). The upregulation, deacetylation, and nuclear localization of SOX9 is associated with tamoxifen resistance in BC (80). In the GEO dataset, SOX9 was upregulated in BM samples of HER2-positive BC. However, SOX9 was downregulated in all the subtypes of BC, and its expression was independent of the HER2 status. Furthermore, HER2-positive BC patients with higher SOX9 expression were associated with poor RFS and OS than those with low SOX9 expression.
FN1 promotes EMT and cancer cell migration (81,82). LUM inhibits MMPs and BC progression, and is co-expressed with COL1A2 in BCBM (83). VCAN is associated with poor OS in BC (84-86). This study showed that the expression levels of FN1, LUM, and VCAN were significantly lower in BM samples of HER2-positive BC than the primary BC patients. However, further verification demonstrated upregulation of FN1, LUM, and VCAN in primary BC tissues. Furthermore, FN1 was highly expressed in HER2-positive MBC compared to the HER2-negative MBC patients. ITGB1, the β1 integrin subunit, mediates initiation, growth, and progression of BC (87). In the present study, ITGB1 was downregulated in primary BC independent of the subtypes and in the HER2-positive BCBM patients.
Several downregulated genes in HER2-positive BCBM patients were upregulated in majority of the BC subtypes and were associated with poorer survival outcomes. This suggested that the tumor microenvironment was different in patients with BM compared to those without BM. Previous studies showed that COL1A2, COL3A1, MMP3, MMP14, MMP2, and FN were downregulated in MBC samples compared to the primary BC samples. These genes are potentially involved in the interactions of the MBC cells with the microenvironment and may be necessary for the metastasis to the lymph nodes and their survival in the new microenvironment (88-91). Another hypothesis is that some of these genes are mainly expressed in the stromal tissues. However, stromal tissue levels are significantly lower in the brain compared to the primary BC. Furthermore, patients with LUM-positive cancer cells were associated with longer survival rates, but patients with LUM-positive stromal tissues were associated with shorter survival rates (92). This suggested different biological roles for some EMT and ECM genes in the stroma and the cancer cells, especially in the context of BM.
Conclusions
In conclusion, 1,056 DEGs (767 downregulated and 289 upregulated) and 14 hub genes were identified in BM samples of HER2-positive BC. Furthermore, CD44, COL1A2, MMP14, POSTN, and SOX9 were associated with BM in BC and survival outcomes of patients with HER2-positive BC. These genes played a significant role in the metastatic tumor microenvironment. However, as present results are based on a single database, bias is existed in current investigation. In addition, above hub genes are components of the stromal compartment which are required for normal tissue homeostasis. Although it has been known that the homeostasis is re-established during BM, it is still difficult to find proper way to keep balance between BM management and tissue homeostasis when targeting the hub genes. Further studies are still required to decipher the biological roles and mechanisms of these hub genes in BM of HER2-positive BC patients.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2715/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2715/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- IARC. Latest global cancer data: cancer burden rises to 19.3 million new cases and 10.0 million cancer deaths in 2020. 2021. Available online: https://www.iarc.fr/faq/latest-global-cancer-data-2020-qa/
- Brosnan EM, Anders CK. Understanding patterns of brain metastasis in breast cancer and designing rational therapeutic strategies. Ann Transl Med 2018;6:163. [Crossref] [PubMed]
- Pasquier D, Darlix A, Louvel G, et al. Treatment and outcomes in patients with central nervous system metastases from breast cancer in the real-life ESME MBC cohort. Eur J Cancer 2020;125:22-30. [Crossref] [PubMed]
- Kromer C, Xu J, Ostrom QT, et al. Estimating the annual frequency of synchronous brain metastasis in the United States 2010-2013: a population-based study. J Neurooncol 2017;134:55-64. [Crossref] [PubMed]
- Soffietti R, Abacioglu U, Baumert B, et al. Diagnosis and treatment of brain metastases from solid tumors: guidelines from the European Association of Neuro-Oncology (EANO). Neuro Oncol 2017;19:162-74. [Crossref] [PubMed]
- Dawood S, Ueno NT, Valero V, et al. Incidence of and survival following brain metastases among women with inflammatory breast cancer. Ann Oncol 2010;21:2348-55. [Crossref] [PubMed]
- Warren LE, Guo H, Regan MM, et al. Inflammatory breast cancer and development of brain metastases: risk factors and outcomes. Breast Cancer Res Treat 2015;151:225-32. [Crossref] [PubMed]
- Lin NU, Vanderplas A, Hughes ME, et al. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the National Comprehensive Cancer Network. Cancer 2012;118:5463-72. [Crossref] [PubMed]
- Pestalozzi BC, Holmes E, de Azambuja E, et al. CNS relapses in patients with HER2-positive early breast cancer who have and have not received adjuvant trastuzumab: a retrospective substudy of the HERA trial (BIG 1-01). Lancet Oncol 2013;14:244-8. [Crossref] [PubMed]
- Lin NU, Claus E, Sohl J, et al. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: high incidence of central nervous system metastases. Cancer 2008;113:2638-45. [Crossref] [PubMed]
- Kennecke H, Yerushalmi R, Woods R, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol 2010;28:3271-7. [Crossref] [PubMed]
- Brastianos PK, Curry WT, Oh KS. Clinical discussion and review of the management of brain metastases. J Natl Compr Canc Netw 2013;11:1153-64. [Crossref] [PubMed]
- Lu-Emerson C, Eichler AF. Brain metastases. Continuum (Minneap Minn) 2012;18:295-311. [Crossref] [PubMed]
- Sperduto PW, Kased N, Roberge D, et al. Summary report on the graded prognostic assessment: an accurate and facile diagnosis-specific tool to estimate survival for patients with brain metastases. J Clin Oncol 2012;30:419-25. [Crossref] [PubMed]
- Komorowski AS, Warner E, MacKay HJ, et al. Incidence of Brain Metastases in Nonmetastatic and Metastatic Breast Cancer: Is There a Role for Screening? Clin Breast Cancer 2020;20:e54-64. [Crossref] [PubMed]
- Bartsch R, Wenzel C, Steger GG. Trastuzumab in the management of early and advanced stage breast cancer. Biologics 2007;1:19-31. [PubMed]
- Van Poznak CH, Temin S, Yee GC, et al. American Society of Clinical Oncology executive summary of the clinical practice guideline update on the role of bone-modifying agents in metastatic breast cancer. J Clin Oncol 2011;29:1221-7. [Crossref] [PubMed]
- Bendell JC, Domchek SM, Burstein HJ, et al. Central nervous system metastases in women who receive trastuzumab-based therapy for metastatic breast carcinoma. Cancer 2003;97:2972-7. [Crossref] [PubMed]
- Brastianos PK, Carter SL, Santagata S, et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov 2015;5:1164-77. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- McMullin RP, Wittner BS, Yang C, et al. A BRCA1 deficient-like signature is enriched in breast cancer brain metastases and predicts DNA damage-induced poly (ADP-ribose) polymerase inhibitor sensitivity. Breast Cancer Res 2014;16:R25. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M. The KEGG database. Novartis Found Symp 2002;247:91-101; discussion 101-3, 119-28, 244-52.
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808-15. [Crossref] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson 2012;14:83. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022;25:18-27. [Crossref] [PubMed]
- Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009;459:1005-9. [Crossref] [PubMed]
- Saunus JM, Quinn MC, Patch AM, et al. Integrated genomic and transcriptomic analysis of human brain metastases identifies alterations of potential clinical significance. J Pathol 2015;237:363-78. [Crossref] [PubMed]
- Kuroiwa Y, Nakayama J, Adachi C, et al. Proliferative Classification of Intracranially Injected HER2-positive Breast Cancer Cell Lines. Cancers (Basel) 2020;12:1811. [Crossref] [PubMed]
- Lu X, Gao C, Liu C, et al. Identification of the key pathways and genes involved in HER2-positive breast cancer with brain metastasis. Pathol Res Pract 2019;215:152475. [Crossref] [PubMed]
- Waugh DJJ, McClatchey A, Montgomery N, et al. Adhesion and penetration: Two sides of CD44 signal transduction cascades in the context of cancer cell metastasis. In: Stern R. editor. Hyaluronan in Cancer Biology. Cambridge: Academic Press, 2009:109-25.
- Al-Othman N, Alhendi A, Ihbaisha M, et al. Role of CD44 in breast cancer. Breast Dis 2020;39:1-13. [Crossref] [PubMed]
- Hassn Mesrati M, Syafruddin SE, Mohtar MA, et al. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules 2021;11:1850. [Crossref] [PubMed]
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell 2011;147:275-92. [Crossref] [PubMed]
- Fares J, Fares MY, Khachfe HH, et al. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther 2020;5:28. [Crossref] [PubMed]
- Barraza DE, Zampini R, Apichela SA, et al. Modifications of extracellular matrix features in the left and right uterine horns during the embryo pre-implantation period in Vicugna pacos. Theriogenology 2020;157:440-8. [Crossref] [PubMed]
- Folgueira MA, Maistro S, Katayama ML, et al. Markers of breast cancer stromal fibroblasts in the primary tumour site associated with lymph node metastasis: a systematic review including our case series. Biosci Rep 2013;33:e00085. [Crossref] [PubMed]
- Shah M, Huang D, Blick T, et al. An MMP13-selective inhibitor delays primary tumor growth and the onset of tumor-associated osteolytic lesions in experimental models of breast cancer. PLoS One 2012;7:e29615. [Crossref] [PubMed]
- Zhang B, Cao X, Liu Y, et al. Tumor-derived matrix metalloproteinase-13 (MMP-13) correlates with poor prognoses of invasive breast cancer. BMC Cancer 2008;8:83. [Crossref] [PubMed]
- Dumortier M, Ladam F, Damour I, et al. ETV4 transcription factor and MMP13 metalloprotease are interplaying actors of breast tumorigenesis. Breast Cancer Res 2018;20:73. [Crossref] [PubMed]
- Pivetta E, Scapolan M, Pecolo M, et al. MMP-13 stimulates osteoclast differentiation and activation in tumour breast bone metastases. Breast Cancer Res 2011;13:R105. [Crossref] [PubMed]
- Sendon-Lago J, Seoane S, Eiro N, et al. Cancer progression by breast tumors with Pit-1-overexpression is blocked by inhibition of metalloproteinase (MMP)-13. Breast Cancer Res 2014;16:505. [Crossref] [PubMed]
- Schulten HJ, Bangash M, Karim S, et al. Comprehensive molecular biomarker identification in breast cancer brain metastases. J Transl Med 2017;15:269. [Crossref] [PubMed]
- Mendes O, Kim HT, Stoica G. Expression of MMP2, MMP9 and MMP3 in breast cancer brain metastasis in a rat model. Clin Exp Metastasis 2005;22:237-46. [Crossref] [PubMed]
- Stark AM, Anuszkiewicz B, Mentlein R, et al. Differential expression of matrix metalloproteinases in brain- and bone-seeking clones of metastatic MDA-MB-231 breast cancer cells. J Neurooncol 2007;81:39-48. [Crossref] [PubMed]
- Li Z, Takino T, Endo Y, et al. Activation of MMP-9 by membrane type-1 MMP/MMP-2 axis stimulates tumor metastasis. Cancer Sci 2017;108:347-53. [Crossref] [PubMed]
- Itoh Y, Seiki M. MT1-MMP: a potent modifier of pericellular microenvironment. J Cell Physiol 2006;206:1-8. [Crossref] [PubMed]
- Zeng C, Lin M, Jin Y, et al. Identification of Key Genes Associated with Brain Metastasis from Breast Cancer: A Bioinformatics Analysis. Med Sci Monit 2022;28:e935071. [Crossref] [PubMed]
- Koga Y, Pelizzola M, Cheng E, et al. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res 2009;19:1462-70. [Crossref] [PubMed]
- Kuivaniemi H, Tromp G. Type III collagen (COL3A1): Gene and protein structure, tissue distribution, and associated diseases. Gene 2019;707:151-71. [Crossref] [PubMed]
- Shin K, Lim A, Zhao C, et al. Hedgehog signaling restrains bladder cancer progression by eliciting stromal production of urothelial differentiation factors. Cancer Cell 2014;26:521-33. [Crossref] [PubMed]
- Shin K, Lee J, Guo N, et al. Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature 2011;472:110-4. [Crossref] [PubMed]
- Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J Surg Oncol 2016;14:297. [Crossref] [PubMed]
- Zhuo C, Li X, Zhuang H, et al. Elevated THBS2, COL1A2, and SPP1 Expression Levels as Predictors of Gastric Cancer Prognosis. Cell Physiol Biochem 2016;40:1316-24. [Crossref] [PubMed]
- Misawa K, Kanazawa T, Misawa Y, et al. Hypermethylation of collagen α2 (I) gene (COL1A2) is an independent predictor of survival in head and neck cancer. Cancer Biomark 2011;10:135-44. [Crossref] [PubMed]
- Yu Y, Liu D, Liu Z, et al. The inhibitory effects of COL1A2 on colorectal cancer cell proliferation, migration, and invasion. J Cancer 2018;9:2953-62. [Crossref] [PubMed]
- Zhang H, Ding C, Li Y, et al. Data mining-based study of collagen type III alpha 1 (COL3A1) prognostic value and immune exploration in pan-cancer. Bioengineered 2021;12:3634-46. [Crossref] [PubMed]
- Wang R, Fu L, Li J, et al. Microarray Analysis for Differentially Expressed Genes Between Stromal and Epithelial Cells in Development and Metastasis of Invasive Breast Cancer. J Comput Biol 2020;27:1631-43. [Crossref] [PubMed]
- Xiong G, Deng L, Zhu J, et al. Prolyl-4-hydroxylase α subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 2014;14:1. [Crossref] [PubMed]
- Yao G, Zhao K, Bao K, et al. Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer. Open Med (Wars) 2022;17:329-40. [Crossref] [PubMed]
- Yousefi M, Dehghani S, Nosrati R, et al. Current insights into the metastasis of epithelial ovarian cancer - hopes and hurdles. Cell Oncol (Dordr) 2020;43:515-38. [Crossref] [PubMed]
- Takeshita S, Kikuno R, Tezuka K, et al. Osteoblast-specific factor 2: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem J 1993;294:271-8. [Crossref] [PubMed]
- González-González L, Alonso J. Periostin: A Matricellular Protein With Multiple Functions in Cancer Development and Progression. Front Oncol 2018;8:225. [Crossref] [PubMed]
- Ratajczak-Wielgomas K, Grzegrzolka J, Piotrowska A, et al. Periostin expression in cancer-associated fibroblasts of invasive ductal breast carcinoma. Oncol Rep 2016;36:2745-54. [Crossref] [PubMed]
- Rachner TD, Göbel A, Hoffmann O, et al. High serum levels of periostin are associated with a poor survival in breast cancer. Breast Cancer Res Treat 2020;180:515-24. [Crossref] [PubMed]
- Zhang Y, Zhang G, Li J, et al. The expression analysis of periostin in human breast cancer. J Surg Res 2010;160:102-6. [Crossref] [PubMed]
- Labrèche C, Cook DP, Abou-Hamad J, et al. Periostin gene expression in neu-positive breast cancer cells is regulated by a FGFR signaling cross talk with TGFβ/PI3K/AKT pathways. Breast Cancer Res 2021;23:107. [Crossref] [PubMed]
- Chen G, Wang Y, Zhao X, et al. A positive feedback loop between Periostin and TGFβ1 induces and maintains the stemness of hepatocellular carcinoma cells via AP-2α activation. J Exp Clin Cancer Res 2021;40:218. [Crossref] [PubMed]
- Yue H, Li W, Chen R, et al. Stromal POSTN induced by TGF-β1 facilitates the migration and invasion of ovarian cancer. Gynecol Oncol 2021;160:530-8. [Crossref] [PubMed]
- Ruan H, Hu S, Zhang H, et al. Upregulated SOX9 expression indicates worse prognosis in solid tumors: a systematic review and meta-analysis. Oncotarget 2017;8:113163-73. [Crossref] [PubMed]
- Zhang S, Che D, Yang F, et al. Tumor-associated macrophages promote tumor metastasis via the TGF-β/SOX9 axis in non-small cell lung cancer. Oncotarget 2017;8:99801-15. [Crossref] [PubMed]
- Ma Y, Shepherd J, Zhao D, et al. SOX9 is a critical regulator of triple-negative breast cancer growth and invasion. Cancer Res 2018;78:abstr 3347.
- Chakravarty G, Moroz K, Makridakis NM, et al. Prognostic significance of cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. Exp Biol Med (Maywood) 2011;236:145-55. [Crossref] [PubMed]
- Ma Y, Bollu L, Hill J, et al. SOX9 controls TNBC growth and metastasis via regulating the expression of apoptosis and EMT genes. Cancer Res 2019;79:abstr 2618.
- Lei B, Zhang Y, Liu T, et al. Sox9 upregulation in breast cancer is correlated with poor prognosis and the CD44+/CD24−/low phenotype. Int J Clin Exp Pathol 2016;9:7345-51.
- Jeselsohn R, Cornwell M, Pun M, et al. Embryonic transcription factor SOX9 drives breast cancer endocrine resistance. Proc Natl Acad Sci U S A 2017;114:E4482-91. [Crossref] [PubMed]
- Liu Y, Xue M, Du S, et al. Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat Commun 2019;10:1637. [Crossref] [PubMed]
- Graf F, Horn P, Ho AD, et al. The extracellular matrix proteins type I collagen, type III collagen, fibronectin, and laminin 421 stimulate migration of cancer cells. FASEB J 2021;35:e21692. [Crossref] [PubMed]
- Hozhabri H, Ghasemi Dehkohneh RS, Razavi SM, et al. Comparative analysis of protein-protein interaction networks in metastatic breast cancer. PLoS One 2022;17:e0260584. [Crossref] [PubMed]
- Kischel P, Waltregny D, Dumont B, et al. Versican overexpression in human breast cancer lesions: known and new isoforms for stromal tumor targeting. Int J Cancer 2010;126:640-50. [Crossref] [PubMed]
- Suwiwat S, Ricciardelli C, Tammi R, et al. Expression of extracellular matrix components versican, chondroitin sulfate, tenascin, and hyaluronan, and their association with disease outcome in node-negative breast cancer. Clin Cancer Res 2004;10:2491-8. [Crossref] [PubMed]
- Ricciardelli C, Brooks JH, Suwiwat S, et al. Regulation of stromal versican expression by breast cancer cells and importance to relapse-free survival in patients with node-negative primary breast cancer. Clin Cancer Res 2002;8:1054-60. [PubMed]
- Park CC, Zhang H, Pallavicini M, et al. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res 2006;66:1526-35. [Crossref] [PubMed]
- Srour MK, Gao B, Dadmanesh F, et al. Gene expression comparison between primary triple-negative breast cancer and paired axillary and sentinel lymph node metastasis. Breast J 2020;26:904-10. [Crossref] [PubMed]
- Hao X, Sun B, Hu L, et al. Differential gene and protein expression in primary breast malignancies and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. Cancer 2004;100:1110-22. [Crossref] [PubMed]
- Suzuki M, Tarin D. Gene expression profiling of human lymph node metastases and matched primary breast carcinomas: clinical implications. Mol Oncol 2007;1:172-80. [Crossref] [PubMed]
- Feng Y, Sun B, Li X, et al. Differentially expressed genes between primary cancer and paired lymph node metastases predict clinical outcome of node-positive breast cancer patients. Breast Cancer Res Treat 2007;103:319-29. [Crossref] [PubMed]
- Ishiwata T, Cho K, Kawahara K, et al. Role of lumican in cancer cells and adjacent stromal tissues in human pancreatic cancer. Oncol Rep 2007;18:537-43. [Crossref] [PubMed]


