
Sarcomatoid renal cell carcinoma: genomic insights from sequencing of matched sarcomatous and carcinomatous components
Introduction
The estimated new kidney cancer cases diagnosed each year in the United Sates and in the world are ~63,000 and ~300,000, respectively (1,2). Renal cell carcinoma (RCC) represents over 90% of kidney cancer and consists of a group of malignancies arising from the renal epithelium and exhibiting distinct histopathological features (3-6). The 2004 WHO classification listed 12 different subtypes of RCC (4). With a better understanding of the molecular pathogenesis of RCC, the 2013 International Society of Urological Pathology (ISUP) consensus conference added several new entities (6). Major RCC subtypes are clear cell RCC (ccRCC) (~75%), papillary RCC (pRCC) (~15%), chromophobe RCC (chRCC) (~5%), and unclassified RCC (uRCC) (4–6%) (6,7). Large-scale genomics of major RCC subtypes led by The Cancer Genome Atlas (TCGA) have been reported, which delineate the genomic landscapes of ccRCC (KIRC), pRCC (KIRP), and chRCC (KICH) (8-11). Furthermore, subsequent studies have begun to elucidate the prognostic and predictive values of prevalent mutations in ccRCC, which is likely to impact clinical management of kidney cancer patients in the near future (12-18).
Sarcomatoid components can be detected in various epithelial malignancies, featuring morphological characteristics typical of a sarcoma and implicating an underlying epithelial-mesenchymal transition (EMT) (19,20). Sarcomatoid components can arise in all subtypes of RCC (21) but with higher incidences in ccRCC and chRCC (22,23). Immunohistochemical and genetic studies indicated that sarcomatoid RCC (sRCC) does not develop de novo but results from transformation/differentiation/dedifferentiation of pre-existing RCC (21,24,25). Hence sRCC does not represent a distinct subtype and is classified according underlying histology; when no epithelial component is present, these tumors are categorized as uRCC. In general, sRCC is associated with an aggressive clinical course and portends a poor therapeutic outcome (22,26-28). Furthermore, increasing percentages of sarcomatoid component within individual RCCs are associated with worsening outcome and carry prognostic values (26,29,30). Accordingly, a better understanding of underlying molecular pathology is of paramount significance.
Genomics of sRCC
Several of the previously reported studies that have examined the genomic aberrations present in ccRCC and chRCC have included patients with sarcomatoid histology (8,10,31). However, interpreting differences in the molecular biology of patients with sRCC in these studies is difficult due to several methodological issues (e.g., different platforms for sequencing, mixed cohorts, small overall numbers). Complicating the issue further is the presence of intratumor heterogeneity and the fact that even on a single slide of paraffin-embedded tissue there can be both areas of sRCC mixed with pure ccRCC and normal renal epithelium. As one could imagine, DNA extracted from these samples would be derived from various sources. While parsing sequencing results for tumor versus normal epithelium can be done quite easily, the segregation of DNA from sRCC and ccRCC is not so simple.
Before the advent of next generation sequencing technology, studies directly comparing matched sarcomatous and carcinomatous components of ccRCC include assessing the mutation status of TP53 and H-RAS (32), determining pattern of allelic loss (25), and immunohistochemistry of EMT markers (20). In an effort to better elucidate the genomic aberrations present in these tumors, two groups recently reported on their dedicated studies of sRCC tumors. Bi et al. reported their findings in Proceedings of the National Academy of Sciences of the United States of America and Malouf et al. published their study in European Urology, both of which were made available in February 2016 (33,34). Both groups should be commended for their efforts to further describe this aggressive and frequently lethal tumor variant.
While the goals of both studies were the same—identify the genomic alterations in sRCC—the design and approach of the studies differed. Bi et al. dedicated their study to tumors with sRCC occurring in the presence of clear cell histology, sarcomatoid clear cell RCC (sccRCC). Malouf et al. included both tumors with sccRCC and tumors with sarcomatoid elements occurring in conjunction with varying histologies (e.g., papillary, unclassified, collecting duct). Both of these studies provided excellent details on the molecular aberrations specific to sRCC and the results of these studies present a number of interesting observations that will surely impact future research.
A cohort of 21 tumors with sequencing results of sufficient quality was initially included in the Bi et al. study. Each tumor had matched normal, carcinomatous, and sarcomatoid elements (microdissected), submitted for whole-exome sequencing. The mean depth of independent reads was 135, 177, and 171 for normal, carcinomatous, and sarcomatoid elements, respectively. Two of the 21 matched samples were found to have significantly higher somatic single nucleotide variants (SSNVs) in their matched tumor elements. One of these tumors had a mutation in mutS homolog 2 (MSH2), and the other had a mutation in polymerase ε (POLE). Their mutational signatures were consistent with mismatch repair deficiency, and they were excluded from the grouped analysis of the other 19 tumors. In the 19 matched tumors samples analyzed, Bi et al. found 41.7% (45/108) of the SSNVs were shared among the matched carcinomatous and sarcomatoid elements. Most of these shared SSNVs were within genes commonly mutated in ccRCC (e.g., VHL, PBRM1, and SETD2). Among these tumors the sarcomatoid elements were found to have a significantly higher average mutational burden (45 vs. 18 SSVNs) and nearly twice the length of loss of heterozygosity (LOH) events (913 vs. 460 Mb), which was also statistically significant. They also found that the sarcomatoid elements had significantly more frequent alterations occurring in known cancer genes. The most frequently mutated among these genes in the sarcomatoid element was tumor protein p53 (TP53). They reported 6 out of 19 (31.6%) sarcomatoid elements had TP53 mutations compared to zero in the matched carcinomatous elements. Bi et al. also highlighted sarcomatoid-specific mutations in BRACA1 associated protein 1 (BAP1) in 2/19 (10.5%) and AT-rich interaction domain 1A (ARID1A) in 3/19 (15.7%) samples. They also observed that mutations in TP53, BAP1, and ARID1A were all mutually exclusive among the 19 tumors. Sarcomatoid elements were also found to have more frequent LOH events among chromosomes 1p, 9, 10, 17p, 18 and 22. Ito et al. reported a similar enrichment for such copy number events in sRCC (35). Lastly, Bi et al. presented several novel SSNVs that have not regularly been associated with RCC which were found to be more common or exclusive to the sarcomatoid elements among the 19 tumors. This included alterations in; FAT atypical cadherin 1 (FAT1), FAT2, FAT3, tumor susceptibility 101 (TSG101), ligand dependent nuclear receptor interacting factor 1 (LRIF1), required for cell differentiation 1 homolog (RQCD1), and protein tyrosine kinase 7 (PTK7).
The study published by Malouf et al. included essentially three different cohorts. The first cohort was similar to the 19-tumor cohort presented by Bi et al., and it included three tumors with paired clear cell (carcinomatous) and sarcomatoid elements after microdissection. This cohort underwent targeted sequencing of both matched elements using a custom panel of 236 frequently mutated cancer-related genes and 37 introns frequently rearranged in cancer (average exon coverage of 819x). Of note this panel did not include the genes FAT1, FAT2, FAT3, TSG101, LRIF1, RQCD1, or PTK7. Also, they did not report using matched normal tissue from any patients in their targeted sequencing analysis, which likely limits the interpretation of copy number aberrations for these tumors. The sequencing results of this cohort stand somewhat in contrast to the results of the study above. Malouf et al. reported identical alterations in two of the three matched samples (i.e., exact same type and number of alterations in both clear cell and sarcomatoid elements). In the third sample they saw similar homozygous deletions in VHL but found multiple distinct inactivating mutations in TP53 and phosphatase and tensin homolog (PTEN) that differed between the two elements. The third case also had a unique amplification of Janus kinase 2 (JAK2) in the sarcomatoid element, which, when taken together with the TP53 and PTEN mutations, may suggest a divergent course of evolution for this tumor. In the second cohort, they analyzed 23 tumors with sRCC arising from a mixture of carcinomatous backgrounds including clear cell, unclassified, collecting duct, papillary, and mucinous tubular and spindle cell carcinoma. Most of these tumors were primary kidney specimens (88.5%), except for three which were from metastatic sites (peritoneal nodule, lymph node, and liver). In this cohort, they found TP53 to be the most frequently altered gene (11/23, 42.3%). They also reported a relatively high number of cyclin-dependent kinase inhibitor 2A (CDKN2A; 7/23, 26.9%) and neurofibromin 2 (NF2; 5/23, 19.2%) alterations among these tumors. In their third cohort, the investigators employed whole-exome sequencing on four tumors with sccRCC, not microdissected. They reported a lower overall median mutation rate in these four cases (37.5 mutations) compared to the median rate in TCGA (49 mutations) for ccRCC (8). In two of these four cases they went on to test multiple regions from the primary tumors (4 regions in one and two in the other) to evaluate intratumoral heterogeneity using Sanger sequencing for VHL and TP53 genes only. For these two cases they report no finding of intratumor heterogeneity in regards to these two genes.
Integrating the results of these studies helps us answer several questions about the molecular framework of sRCC. First the truncal events and shared genomic aberrations between both the carcinomatous elements and sarcomatoid element seen in both studies confirm that sRCC arises from RCC. Next, the notion that the sarcomatoid element represents a dedifferentiated progression of RCC is supported by the increased overall mutational burden and copy number aberrations seen in the sarcomatoid elements compared to the carcinomatous elements from Bi et al. The increase in aberrations of known cancer genes (TP53, NF2, CDKN2A) also supports the sentiment that the sarcomatoid elements are driving pathogenesis in these tumors. Oda et al. published a study in 1995 reporting a mutation rate of 78.6% (11 of 14) for TP53 in the sarcomatoid elements of sRCC tumors using polymerase chain reaction (32). The carcinomatous elements, or background histology, for this cohort included both mixed and granular subtypes, somewhat limiting the application of these results. While Bi et al. clearly show an enrichment of TP53 aberrations (31.6%) in the sarcomatoid elements among primary ccRCC tumors, caution must be used when interpreting the even more enriched results (42.3%) from the 26 sRCC tumors reported in the Malouf et al. study. The latter study included diverse primary RCC histologies and also included metastatic tumors, which previously have been shown to be enriched for TP53 aberrations irrespective of sarcomatoid features (36). Similarly the finding of increased NF2 mutations occurring in sRCC may also be limited due to the diverse background of primary RCC histologies in this cohort. Our understanding of the molecular composition and the clinical implications of uRCC are both poorly defined and poorly understood. As a significant number of the NF2 and TP53 aberrations occurred in these unclassified tumors, attributing the results to sRCC may be problematic. Another interesting finding is the identification of the two tumors from Bi et al. with mutational signatures consistent with mismatch repair deficiency. Tumors such as these, and maybe even sarcomatoid variants in general, may derive significant benefit from immune checkpoint blockade in the treatment of metastatic disease (37,38). A summary of some of the differences among the tumor cohorts analyzed in these studies can be found in Table 1.
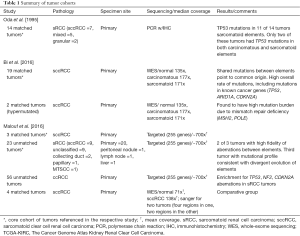
Full table
Both of these studies are novel in their attempt to better understand this very clinically relevant and aggressive disease. However, sRCC is a relatively rare entity, and both studies have small cohorts, which may hinder their generalizability. The rarity of this disease also exposes both studies to significant selection bias. This may include selection of tumors for analysis with the most tissue available (large tumors), those with the most aggressive course (likely to have been sequenced), and likely other confounding variables. The use of different sequencing platforms and the mix of histologies in the Malouf et al. study make pooling and comparing of the results difficult. While the genomic underpinnings of sRCC in approximately 1/3 of patients may be explained by the results of these studies (i.e., TP53, NF2, CDKN2A aberrations), there is still no clear molecular explanation of sRCC development in the majority of cases.
Conclusions
Both Bi et al. and Malouf et al. have conducted and published novel genomic studies of renal tumors with sarcomatoid variant histology. The results have definitively demonstrated that progressive dedifferentiation is the source of the sarcomatoid elements in RCC. They have also identified key genomic aberrations (e.g., TP53, CDKN2A, copy number changes) present in sRCC that may explain its aggressive clinical course and may become potential targets for therapy. We hope future research efforts build upon this work to pursue better treatment and management strategies for patients with this disease.
Acknowledgments
Funding: Supported by the Sidney Kimmel Center for Prostate and Urologic Cancers, the NIH/NCI Cancer Center Support Grant P30 CA008748, the Ruth L. Kirschstein National Research Service Award T32CA082088 (BJM), the Jill and Jeffrey Weiss Fund to the Cure of Kidney Cancer, and the J. Randall & Kathleen L. MacDonald Kidney Cancer Research Fund (JJH).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Peng Zhang (Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.07.30). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Global Burden of Disease Cancer Collaboration. The Global Burden of Cancer 2013. JAMA Oncol 2015;1:505-27. [Crossref] [PubMed]
- Kovacs G, Akhtar M, Beckwith BJ, et al. The Heidelberg classification of renal cell tumours. J Pathol 1997;183:131-3. [Crossref] [PubMed]
- Eble JN SG, Epstein JI, Sesterhenn IA. Pathology and genetics of tumours of the urinary system and male genital organs: Lyon: IARC Press, 2004.
- Lopez-Beltran A, Scarpelli M, Montironi R, et al. 2004 WHO classification of the renal tumors of the adults. Eur Urol 2006;49:798-805. [Crossref] [PubMed]
- Srigley JR, Delahunt B, Eble JN, et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am J Surg Pathol 2013;37:1469-89. [Crossref] [PubMed]
- Sankin A, Hakimi AA, Hsieh JJ, et al. Metastatic non-clear cell renal cell carcinoma: an evidence based review of current treatment strategies. Front Oncol 2015;5:67. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43-9. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 2016;374:135-45. [Crossref] [PubMed]
- Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014;26:319-30. [Crossref] [PubMed]
- Chen F, Zhang Y, Şenbabaoğlu Y, et al. Multilevel Genomics-Based Taxonomy of Renal Cell Carcinoma. Cell Rep 2016;14:2476-89. [Crossref] [PubMed]
- Hakimi AA, Chen YB, Wren J, et al. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. Eur Urol 2013;63:848-54. [Crossref] [PubMed]
- Hakimi AA, Ostrovnaya I, Reva B, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res 2013;19:3259-67. [Crossref] [PubMed]
- Kapur P, Peña-Llopis S, Christie A, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol 2013;14:159-67. [Crossref] [PubMed]
- Hsieh J, Chen D, Wang P, et al. Identification of efficacy biomarkers in a large metastatic renal cell carcinoma (mRCC) cohort through next generation sequencing (NGS): Results from RECORD-3. J Clin Oncol 2015;33:abstr 4509.
- Wei EY, Hsieh JJ. A river model to map convergent cancer evolution and guide therapy in RCC. Nat Rev Urol 2015;12:706-12. [Crossref] [PubMed]
- Wei EY, Chen YB, Hsieh JJ. Genomic characterisation of two cancers of unknown primary cases supports a kidney cancer origin. BMJ Case Rep 2015;2015. pii: bcr2015212685.
- Sankin A, Hakimi AA, Mikkilineni N, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med 2014;3:1485-92. [Crossref] [PubMed]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178-96. [Crossref] [PubMed]
- Conant JL, Peng Z, Evans MF, et al. Sarcomatoid renal cell carcinoma is an example of epithelial--mesenchymal transition. J Clin Pathol 2011;64:1088-92. [Crossref] [PubMed]
- Störkel S, Eble JN, Adlakha K, et al. Classification of renal cell carcinoma: Workgroup No. 1. Union Internationale Contre le Cancer (UICC) and the American Joint Committee on Cancer (AJCC). Cancer 1997;80:987-9. [Crossref] [PubMed]
- Cheville JC, Lohse CM, Zincke H, et al. Sarcomatoid renal cell carcinoma: an examination of underlying histologic subtype and an analysis of associations with patient outcome. Am J Surg Pathol 2004;28:435-41. [Crossref] [PubMed]
- Shuch B, Bratslavsky G, Linehan WM, et al. Sarcomatoid renal cell carcinoma: a comprehensive review of the biology and current treatment strategies. Oncologist 2012;17:46-54. [Crossref] [PubMed]
- Delahunt B. Sarcomatoid renal carcinoma: the final common dedifferentiation pathway of renal epithelial malignancies. Pathology 1999;31:185-90. [Crossref] [PubMed]
- Jones TD, Eble JN, Wang M, et al. Clonal divergence and genetic heterogeneity in clear cell renal cell carcinomas with sarcomatoid transformation. Cancer 2005;104:1195-203. [Crossref] [PubMed]
- Golshayan AR, George S, Heng DY, et al. Metastatic sarcomatoid renal cell carcinoma treated with vascular endothelial growth factor-targeted therapy. J Clin Oncol 2009;27:235-41. [Crossref] [PubMed]
- Molina AM, Tickoo SK, Ishill N, et al. Sarcomatoid-variant renal cell carcinoma: treatment outcome and survival in advanced disease. Am J Clin Oncol 2011;34:454-9. [Crossref] [PubMed]
- Kyriakopoulos CE, Chittoria N, Choueiri TK, et al. Outcome of patients with metastatic sarcomatoid renal cell carcinoma: results from the International Metastatic Renal Cell Carcinoma Database Consortium. Clin Genitourin Cancer 2015;13:e79-85. [Crossref] [PubMed]
- Zhang BY, Thompson RH, Lohse CM, et al. A novel prognostic model for patients with sarcomatoid renal cell carcinoma. BJU Int 2015;115:405-11. [Crossref] [PubMed]
- Adibi M, Thomas AZ, Borregales LD, et al. Percentage of sarcomatoid component as a prognostic indicator for survival in renal cell carcinoma with sarcomatoid dedifferentiation. Urol Oncol 2015;33:427.e17-23. [Crossref] [PubMed]
- Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013;45:860-7. [Crossref] [PubMed]
- Oda H, Nakatsuru Y, Ishikawa T. Mutations of the p53 gene and p53 protein overexpression are associated with sarcomatoid transformation in renal cell carcinomas. Cancer Res 1995;55:658-62. [PubMed]
- Bi M, Zhao S, Said JW, et al. Genomic characterization of sarcomatoid transformation in clear cell renal cell carcinoma. Proc Natl Acad Sci U S A 2016;113:2170-5. [Crossref] [PubMed]
- Malouf GG, Ali SM, Wang K, et al. Genomic Characterization of Renal Cell Carcinoma with Sarcomatoid Dedifferentiation Pinpoints Recurrent Genomic Alterations. Eur Urol 2016;70:348-57. [Crossref] [PubMed]
- Ito T, Pei J, Dulaimi E, et al. Genomic Copy Number Alterations in Renal Cell Carcinoma with Sarcomatoid Features. J Urol 2016;195:852-8. [Crossref] [PubMed]
- Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225-33. [Crossref] [PubMed]
- Joseph RW, Millis SZ, Carballido EM, et al. PD-1 and PD-L1 Expression in Renal Cell Carcinoma with Sarcomatoid Differentiation. Cancer Immunol Res 2015;3:1303-7. [Crossref] [PubMed]
- Geynisman DM. Anti-programmed Cell Death Protein 1 (PD-1) Antibody Nivolumab Leads to a Dramatic and Rapid Response in Papillary Renal Cell Carcinoma with Sarcomatoid and Rhabdoid Features. Eur Urol 2015;68:912-4. [Crossref] [PubMed]


