
Sweetening of glutamine metabolism in cancer cells by Rho GTPases through convergence of multiple oncogenic signaling pathways
More than 60 years ago, Otto Warburg showed that cancer cells exhibit enhanced glycolysis accompanied by greatly elevated levels of lactate secretion, even in the presence of normal levels of oxygen (1). Warburg suggested that cancer cells arise from normal cells in a two-phase process: phase 1 is “injury” to the respiratory machinery (i.e., mitochondria), followed in phase 2 by enhanced “fermentation” (i.e., production of lactate from glucose) in the protoplasm (i.e., cytosol) (1). Conversion of glucose to lactate as a source of ATP is very inefficient compared to that obtained by complete oxidation of glucose to CO2 via tight coupling of glycolysis to the mitochondrial TCA cycle. Nevertheless, Warburg suggested that given ready access to adequate circulating glucose “fermentation” can provide quiescent cancer cells with the necessary energy requirements (1). For many years the Warburg effect was treated mainly as just another interesting biochemical phenomenon. However, within the last decade the “Warburg effect” has become the subject of intense investigation. We now know that the mitochondria in many cancer cells are not grossly defective as originally envisaged by Warburg, but are metabolically reprogrammed (2). In this case, the carbon of certain metabolites can enter the TCA cycle (anaplerosis) as both an energy source and as a source of intracellular components (e.g., for lipids, nucleic acids, proteins) necessary for rapidly dividing cells. An especially important metabolite in this regard is glutamine (3). Rapidly dividing cancer cells that require substantial amounts of glutamine are said to exhibit “glutamine addiction”. Glutamine is readily converted to the TCA cycle intermediate α-ketoglutarate (α-KG) while at the same time providing nitrogen for DNA, polyamine and non-essential amino acid synthesis. Glutamine is converted to α-KG in a two-step process. Glutamine is hydrolyzed to glutamate by glutaminase, which in turn is converted to α-KG by the glutamate dehydrogenase reaction or by transamination with a suitable α-keto acid substrate (4).
The importance of the Warburg effect and glutamine metabolism in cancer cells was elegantly demonstrated by DeBerardinis et al. (5). These authors used 13C-glucose coupled to 13C-NMR to show that glioblastoma cells in culture copiously convert glucose to lactate (and, to a lesser extent alanine) in the presence of oxygen. Moreover, using 13C-glutamine the authors showed that glutamine is also an important energy source in these cells (glutaminolysis). In these studies, a substantial portion of glutamine carbon (60%) was shown to be directed toward lactate production. DeBerardinis et al. state “A by-product of this flux is robust NADPH production by malic enzyme. The glutaminolytic flux was at least as high as the G6PDH flux, and appeared to be higher than that needed for fatty acid synthesis. This could mean that NADPH generated during glutaminolysis also supplies other anabolic processes such as nucleotide biosynthesis.” Interestingly, glutamine nitrogen was lost to the medium not only as alanine but also as ammonia. Apparently, although glutamine is a source of nitrogen for many compounds in the glioblastoma cells the utilization of glutamine carbon through the TCA cycle provides nitrogen in excess of that needed for these biosynthetic reactions and is excreted in the form of alanine and ammonia (5).
A crucial enzyme in the glutaminolysis pathway is glutaminase (6). In mammals, GLS and GLS2 genes encode two glutaminase isozymes. GLS encodes a kidney-type glutaminase (GLS) and a splice variant [glutaminase C (GAC)]. GLS2 encodes a liver-type glutaminase (GLS2) and a splice variant [glutaminase B (GAB)] (7). Thus, it is not surprising that several cellular oncogenes and their signaling pathways regulate glutaminase activity in cancer cells. For example, many cancer cells that express high levels of c-Myc exhibit elevated glutaminase activity (8), which is associated with de-repression of glutaminase by the microRNA mir23a/b (9). Thus, a molecular link between an oncogenic signal and elevated glutaminase activity has been established. However, this link is not unique to cancer cells because several non-cancerous diseases, such as pulmonary hypertension are also associated with increased glutamine metabolism in the affected cells (10). Because of the unique role of glutaminase in disease processes, intense efforts have been directed toward the development of selective and potent glutaminase inhibitors.
One such glutaminase inhibitor was identified by Wang et al. as a brominated phenanthridinone derivative (compound 968), which was discovered during a chemical screen for inhibitors of a class of oncogenic signaling proteins, namely Rho GTPases (11). In this study, the compound was found to: (I) block oncogenic transformation induced by a well-known guanine nucleotide exchange factor in fibroblasts; and (II) block the growth of human breast cancer and B lymphoma cells, without affecting normal cells (11). A biotinylated derivative of compound 968, when used in a streptavidin pull-down assay revealed that its target protein is the mitochondrial glutaminase (GLS) (11). Additional studies by Wang et al. showed specific down-regulation of GLS by siRNA is associated with suppression of colony formation of cells that constitutively express the oncogenic Rho GTPase family member diffuse B-cell lymphoma (dbl) (11). Further studies revealed that glutaminase inhibition strongly suppresses breast cancer cell proliferation and confirmed a molecular link between elevated glutaminase activity and oncogenic transformation of cells that express Rho GTPase family members (11). The increase in glutaminase activity in these cells was also found to depend on the expression of NF-κB (11). Interestingly, these studies also revealed that the inhibitory effect of the compound 968 could be partially reversed by the supplementation with α-KG, a metabolite in the TCA cycle downstream of GLS activity. Thus, these studies established the importance of glutaminase activity in cancer cells along with its importance in the anaplerotic provision of the TCA cycle intermediate α-KG in cancer cell growth.
A follow-up of the study by Wang et al. (11) by the same group (and the subject of the current commentary) provides a mechanistic link between the action of Rho GTPases and the elevation of glutaminase activity (7). In these studies another potent inhibitor of GLS/GAC was used, namely bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). In this study, Lukey et al. concluded that: (I) the oncogenic transcription factor c-Jun is responsible for the elevation of glutaminase activity by Rho GTPase family members; (II) c-Jun is an important regulator of GLS expression in human breast cancer; and (III) overexpression of this cellular proto-oncogene is sufficient to sensitize breast cancer cells to glutaminase-targeted therapy. Thus, these studies are further confirmation of the importance of glutaminase activity as an important step in providing anaplerotic α-KG, critical for cancer cell growth.
These findings are significant for the following reasons: first, they highlight the contribution of the protein product of the oncogene c-JUN, (which is overexpressed and/or stabilized in many cancers, including breast cancer) in the metabolic rewiring of cancers and provide a molecular explanation for the increased dependency on glutamine metabolism. Second, they offer an explanation for the coordinated metabolic reprogramming and signaling that is required for concomitant increases in cell proliferation and biomass. Finally, elevated expression of GLS in triple negative breast cancer (TNBC), as opposed to estrogen receptor positive (ER+) breast cancer, is now known to be associated with an increase in the protein levels of both proto-oncogenes c-JUN and c-MYC, which transcriptionally upregulate GLS mRNA (12,13). Thus, targeting GLS in TNBC is the basis for several currently ongoing clinical trials.
Despite advances in our understanding of the overall importance of glutaminase activity in cancer metabolism, there are still some unresolved issues. First, whereas GLS is upregulated by several oncogenes such as c-MYC and c-JUN, GLS2 is upregulated by the tumor suppressor protein p53 (14,15). Although wild type p53 contributes to the upregulation of glutaminase it has the added property of promoting antioxidant activity through the increased production of glutathione, which in turn is associated with a significant reduction in tumor development. Thus, Myc- or Jun-induced glutaminolysis supports cancer cell proliferation, whereas p53-induced glutaminolysis is associated with tumor suppression. Hence, there is a need for the design of: (I) compounds that inhibit GLS but not GLS2; and (II) compounds that activate GLS2. In this respect, strategies that are aimed to rescue mutant p53 (which is expressed in a significant fraction of solid tumors) and restore its normal function (16) gain more significance, namely their role in reprogramming of glutamine metabolism for cancer therapeutic purposes.
Second, the recent report by Lukey et al. emphasizes the role of c-Jun signaling downstream of the Rho GTPase activation in the upregulation of glutaminase activity (7). Earlier work by the same group highlights the importance of NF-κB signaling downstream of the same family of Rho GTPases (11). It is interesting to note that mutated p53 stimulates the mevalonate pathway, which provides the geranylgeranyl pyrophosphate moieties covalently linked to Rho family members for insertion into membranes, contributing to their activation (17). By contrast, as described above, wild type p53 enhances GLS2 expression (14,15). The use of glutamine either in normal cell function or in proliferation mirrors the ability of p53 to induce or suppress reactive oxygen species (ROS) production. More importantly, GLS is upregulated by the cellular oncogene c-MYC, which negatively controls the glutaminase-suppressing microRNA, miR23a/b (9). The cellular oncogene c-SRC, which was the first tyrosine kinase oncogene to be discovered, also regulates glutaminase metabolism indirectly through c-MYC. In a recent study, Jain et al. showed that SRC blockade reduces glucose metabolism and the Warburg effect in breast cancer cells through the inhibition of extracellular signal regulated kinase (ERK)1/2-MNK1-eIF4E-mediated cap-dependent translation of c-MYC and the glucose transporter GLUT1 (18). In addition, Src is known to activate the Rho GTPase Rac1 through the participation of the adaptor oncogene CRKII and p120 catenin (19,20). These observations suggest that Rho GTPases activate the Jun N-terminal kinase (JNK), which, in turn, phosphorylates c-Jun, thus strongly activating its properties as a transcription factor. Moreover, evidence suggests the participation of the proliferation/angiogenesis and metastasis suppressor protein SSeCKS (pronounced Essex) [Src suppressed C-kinase substrate; a member of the myristoylated alanine-rich C-kinase substrate (MARCKS) family] in the control of signaling pathways in oncogenesis (21). We propose a hypothesis that integrates the activities of several oncogenes and the tumor suppressor gene p53 (Figure 1). However, the major tenet of this hypothesis, namely the involvement of SSeCKS or MARCKS family of proteins needs to be verified. MARCKS and related SSeCKS proteins serve as a reversible source of phosphoinositides (particularly PIP2), sequestering them in the plasma membrane and preventing their conversion into PIP3. This sequestration inactivates phosphoinositide-dependent kinase (PDK) thus attenuating PI 3-kinase and Akt (22). SSeCKS, which is also known as AKAP12 or gravin, functions as a tumor and metastasis suppressor (23). Recent studies have shown that the functions of SSeCKS/AKAP12 likely involve its ability to negatively influence specific oncogenic signaling pathways through the scaffolding of key mitogenic mediators such as PKC, PKA, cyclins and calmodulin by an electrostatic switch mechanism acting at the membrane (24). Conversely, loss of SSeCKS activity, either by down-regulated gene expression or by phosphorylation leads to hyperactivated PKC-isoenzymes, which in turn leads to increased invasiveness in prostate cancer cells (25).
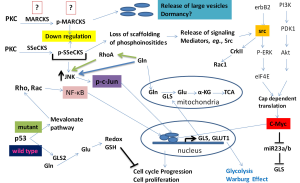
Studies by Gelman and coworkers (23) clearly establish the suppressive role of SSeCKS in Src-induced oncogenesis by regulating the Rho GTPase family members, namely RhoA- and Cdc42-dependent pathways. Since SSeCKS/AKAP12 controls cellular events that cause scaffolding of key signaling molecules such as cyclin D1, calmodulin, PKA, and PKC, this effect would help re-establish the actin-based cytoskeletal architecture (23). Additionally, c-JNK phosphorylation of the MARCKS family member MARCKSL1 determines actin stability, cytoskeletal structure and cell movement, which are important for cancer cell invasiveness, as shown in a prostate cancer model system (26). Working along similar lines, Cohen and coworkers have shown that forced re-expression of the down-regulated SSeCKS reversed the v-Jun induced transformation of 10T1/2 murine fibroblasts (27). These observations suggest that: (I) there is a nexus of regulatory signaling pathways connecting PKC and Rho GTPases to the SSeCKS protein, leading to its down-regulation in cancer; and (II) a down-regulation of SSeCKS is a requirement of v-Jun-induced oncogenic transformation. The oncogenic v-Jun protein differs from c-Jun by two amino-acid substitutions in the C-terminal DNA binding domain and an N-terminal 27 amino acid deletion. Hence, it is very likely that down-regulation of SSeCKS is also an oncogenic pre-requisite for c-Jun-overexpressing cancer cells, including human cancers. Cohen and colleagues (27) have also shown that SSeCKS functions as a tumor suppressor, counteracting the oncogenic effect of Jun as well as Src. Thus, it will be very interesting to determine how Rho GTPase family members that activate JNK negatively influence the expression of MARCKS or related SSeCKS proteins. This may provide a molecular explanation for the mechanism by which Rho GTPases activate JNK, which in turn activates c-JUN specific transcription, providing a molecular link to the studies by Lukey et al. (7). Several pieces of indirect evidence suggest the participation of proteins such as MARCKS in the activation of JNK. For example, one study showed that the MARCKS protein regulates the expression of pro-inflammatory cytokines in murine macrophages through the activation of JNK and NF-κB (28). It is worth noting in this context that SSeCKS, MARCKS and the extracellular matrix protein SPARC are down-regulated in v-Jun transformed cells (29). Thus, it is very likely that in human cancers over-expressing c-Jun, the same genes such as SSeCKS and MARCKS may be targeted for negative regulation. These studies suggest that a molecular relationship may exist between Rho GTPases and elevated JNK signaling (leading to c-JUN activation), and that the MARCKS/SSeCKS family of proteins may provide the link connecting the two (Figure 1). Therefore, it would be very interesting to determine the expression levels of MARCKS/SSeCKS and whether a re-expression or over-expression of MARCKS or SSeCKS negatively influences glutaminase expression as seen in the experimental system employed by Lukey et al. (7).
Third, Lukey et al. have shown that a cell-permeable α-KG precursor partially rescues the growth of dbl-expressing mouse embryonic fibroblasts (MEFs) (7). This raises the immediate concern that cancer cells that reprogram metabolic pathways and elevate intracellular α-KG will negate the therapeutic potential of glutaminase-targeted therapies currently under investigation. Several bypass mechanisms for obtaining anaplerotic carbon are potentially available to the cancer cell and it would be important to investigate their participation before clinically testing any anti-glutaminase therapies.
Fourth, it is interesting to note that an important connection between RhoA-mediated elevation of glutaminase and the generation of large extracellular vesicles has been observed in cancer cells (30). Notably, introduction of specific glutaminase inhibitors (compound 968 or BPTES) strongly inhibits the shedding of these large extracellular vesicles in a variety of cancers such as glioblastoma, metastatic mammary gland adenocarcinoma, pancreatic ductal carcinoma, and pancreatic adenocarcinoma (31).
Thus, it appears that reprogramming of glutamine metabolism and elevated glutaminase activity meet the metabolic demands for increased biomass and lipid synthesis required to replace the plasma membranes lost from these cancer cells as extraverted vesicles. In this respect, it is intriguing that, in a recent study, exosomes from bone marrow mesenchymal cells were shown to contain a microRNA (miR23b) directed against MARCKS mRNA that promotes dormancy in metastatic breast cancer cells (32). Apparently, breast cancer cells fuse with vesicles containing miR23b, inducing suppression of the gene MARCKS. This observation has at least three important implications. First, the miR23b when taken up by the target cancer cells may also down-regulate the expression of GLS, thus preventing the reprogramming of glutamine metabolism, which is crucially needed for cell proliferation and increased metabolic demands. Second, suppressing glutamine metabolism (in the absence of the c-Myc signal) may promote cancer cell dormancy, such as in micrometastases. An increase in GLS may then function as a “wake up” signal for dormant cancer cells to emerge as a proliferating cancer cell. Third, the observation that miR23b also targets MARCKS protein along with GLS strongly supports the hypothesis (Figure 1) that MARCKS/SSeCKS integrates in the Rho GTPases/GLS regulatory mechanism.
Fifth, glutamine is the most abundant free amino acid in the body and is well known to play a regulatory role in several cell specific processes such as apoptosis and cell proliferation (33). Glutamine also has a role in activating metabolism, signal transducing and redox processes, and in stimulating gluconeogenesis, lipogenesis, extracellular matrix production, respiratory burst/cell defenses and heat shock response/chaperone functions (33). Thus, the function of glutamine transcends its classical roles as a simple building block for proteins, a metabolic fuel and a nitrogen source for various biomolecules. Importantly, glutamine plays a role in potentiating the effects of growth factors that facilitate gene expression, cell proliferation and repair (34). In this regard, Rhoads et al. have shown that glutamine activates both extracellular ERK and JNK in intestinal cell proliferation. These proteins are involved in signal transduction pathways stimulated by growth factors, resulting in an increase in AP-1-dependent gene transcription and c-Jun mRNA levels, positively regulating the expression of genes involved in cell division (35). Moreover, glutamine activates the mTORC1 cell proliferation pathway both directly and indirectly through the import of leucine through a bidirectional transport mechanism (36). Therefore, while the studies by Lukey et al. (7) have focused on GLS activity, it is also possible that glutamine per se may have a direct effect in activating JNK under certain cellular circumstances. In consideration of this direct effect, a vicious circle of events may occur wherein glutamine activates JNK, JNK phosphorylates c-Jun, and phosphorylated-c-Jun translocates to the nucleus and activates transcription of GLS. The resultant GLS translocates to the mitochondria where it catalyzes the hydrolysis of glutamine to glutamate and ammonium. Interestingly, a recent report shows that glutamine and its dipeptide analog alanyl-glutamine can increase RhoA expression in intestinal epithelial cells (37). Therefore, future investigations must take into account the participation of glutamine in activating its own metabolism, a phenomenon that could be exploited by cancer cells through Rho GTPases. These observations not only highlight the importance of the studies by Lukey et al. (7) in contributing to our understanding of individual signaling processes involved in the glutamine addiction of cancer cells, but also support the hypothesis that several signaling pathways converge synergistically in this process (Figure 1).
Finally, several very recent reports indicate that the “glutamine addiction” observed in several cancer cell lines in culture is not exactly reproduced in vivo, suggesting that the tumor microenvironment may play an important role in the reprogramming of glutamine metabolism and hence the metabolic phenotype (38,39). Thus, within the core of the tumor, metabolic heterogeneity may exist with respect to glutamine dependency, which may or may not be sensitive to glutaminase-targeted therapies (38-40). This raises question of how the c-Jun activated pathways are regulated by gradients in nutrient concentrations within the highly perfused and poorly perfused regions of the same tumor.
In conclusion, additional studies are needed to evaluate and interpret the therapeutic potential of glutaminase inhibitors, especially within cancers that have reprogrammed glutamine metabolism. Lastly, a better understanding of the Warburg effect is beginning to emerge with regard to the metabolic influences of glutaminase. The paper by Lukey et al. is timely and significant since it focuses on the role of altered oncogenic signaling pathways in the rewiring of cancer cell metabolism and should lay the foundation for personalized and targeted therapy of cancer patients, based on their metabolic phenotype.
Acknowledgments
The authors wish to thank Irwin H. Gelman (Roswell Park Cancer Institute, Buffalo, NY, USA) for his helpful criticisms.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Decai Yu (Hepatobiliary Institute of Nanjing University, Department of Hepatobiliary Surgery, Drum Tower Hospital, The Affiliated Medical School of Nanjing University, Nanjing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.07.43). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Warburg O. On the origin of cancer cells. Science 1956;123:309-14. [Crossref] [PubMed]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012;21:297-308. [Crossref] [PubMed]
- Ahn CS, Metallo CM. Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab 2015;3:1. [Crossref] [PubMed]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011;11:85-95. [Crossref] [PubMed]
- DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A 2007;104:19345-50. [Crossref] [PubMed]
- Szeliga M, Obara-Michlewska M. Glutamine in neoplastic cells: focus on the expression and roles of glutaminases. Neurochem Int 2009;55:71-5. [Crossref] [PubMed]
- Lukey MJ, Greene KS, Erickson JW, et al. The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat Commun 2016;7:11321. [Crossref] [PubMed]
- Dang CV. MYC on the path to cancer. Cell 2012;149:22-35. [Crossref] [PubMed]
- Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009;458:762-5. [Crossref] [PubMed]
- Piao L, Fang YH, Parikh K, et al. Cardiac glutaminolysis: a maladaptive cancer metabolism pathway in the right ventricle in pulmonary hypertension. J Mol Med (Berl) 2013;91:1185-97. [Crossref] [PubMed]
- Wang JB, Erickson JW, Fuji R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 2010;18:207-19. [Crossref] [PubMed]
- Wang X, Chao L, Li X, et al. Elevated expression of phosphorylated c-Jun NH2-terminal kinase in basal-like and "triple-negative" breast cancers. Hum Pathol 2010;41:401-6. [Crossref] [PubMed]
- Horiuchi D, Kusdra L, Huskey NE, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med 2012;209:679-96. [Crossref] [PubMed]
- Hu W, Zhang C, Wu R, et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A 2010;107:7455-60. [Crossref] [PubMed]
- Suzuki S, Tanaka T, Poyurovsky MV, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A 2010;107:7461-6. [Crossref] [PubMed]
- Liu YY. Resuscitating wild-type p53 expression by disrupting ceramide glycosylation: a novel approach to target mutant p53 tumors. Cancer Res 2011;71:6295-9. [Crossref] [PubMed]
- Freed-Pastor WA, Mizuno H, Zhao X, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 2012;148:244-58. [Crossref] [PubMed]
- Jain S, Wang X, Chang CC, et al. Src inhibition blocks c-Myc translation and glucose metabolism to prevent the development of breast cancer. Cancer Res 2015;75:4863-75. [Crossref] [PubMed]
- Liu W, Yue F, Zheng M, et al. The proto-oncogene c-Src and its downstream signaling pathways are inhibited by the metastasis suppressor, NDRG1. Oncotarget 2015;6:8851-74. [Crossref] [PubMed]
- Peglion F, Etienne-Manneville S. p120catenin alteration in cancer and its role in tumour invasion. Philos Trans R Soc Lond B Biol Sci 2013;368:20130015 [Crossref] [PubMed]
- Gelman IH. The role of SSeCKS/gravin/AKAP12 scaffolding proteins in the spaciotemporal control of signaling pathways in oncogenesis and development. Front Biosci 2002;7:d1782-97. [PubMed]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 2005;438:605-11. [Crossref] [PubMed]
- Gelman IH, Gao L. SSeCKS/Gravin/AKAP12 metastasis suppressor inhibits podosome formation via RhoA- and Cdc42-dependent pathways. Mol Cancer Res 2006;4:151-8. [Crossref] [PubMed]
- Su B, Bu Y, Engelberg D, et al. SSeCKS/Gravin/AKAP12 inhibits cancer cell invasiveness and chemotaxis by suppressing a protein kinase C- Raf/MEK/ERK pathway. J Biol Chem 2010;285:4578-86. [Crossref] [PubMed]
- Akakura S, Nochajski P, Gao L, et al. Rb-dependent cellular senescence, multinucleation and susceptibility to oncogenic transformation through PKC scaffolding by SSeCKS/AKAP12. Cell Cycle 2010;9:4656-65. [Crossref] [PubMed]
- Björkblom B, Padzik A, Mohammad H, et al. c-Jun N-terminal kinase phosphorylation of MARCKSL1 determines actin stability and migration in neurons and in cancer cells. Mol Cell Biol 2012;32:3513-26. [Crossref] [PubMed]
- Cohen SB, Waha A, Gelman IH, et al. Expression of a down-regulated target, SSeCKS, reverses v-Jun-induced transformation of 10T1/2 murine fibroblasts. Oncogene 2001;20:141-6. [Crossref] [PubMed]
- Lee SM, Suk K, Lee WH. Myristoylated alanine-rich C kinase substrate (MARCKS) regulates the expression of proinflammatory cytokines in macrophages through activation of p38/JNK MAPK and NF-κB. Cell Immunol 2015;296:115-21. [Crossref] [PubMed]
- Vogt PK. Fortuitous convergences: the beginnings of JUN. Nat Rev Cancer 2002;2:465-9. [Crossref] [PubMed]
- Wilson KF, Erickson JW, Antonyak MA, et al. Rho GTPases and their roles in cancer metabolism. Trends Mol Med 2013;19:74-82. [Crossref] [PubMed]
- Santana SM, Antonyak MA, Cerione RA, et al. Cancerous epithelial cell lines shed extracellular vesicles with a bimodal size distribution that is sensitive to glutamine inhibition. Phys Biol 2014;11:065001 [Crossref] [PubMed]
- Ono M, Kosaka N, Tominaga N, et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci Signal 2014;7:ra63. [Crossref] [PubMed]
- Curi R, Lagranha CJ, Doi SQ, et al. Molecular mechanisms of glutamine action. J Cell Physiol 2005;204:392-401. [Crossref] [PubMed]
- Matés JM, Pérez-Gómez C, Núñez de Castro I, et al. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. Int J Biochem Cell Biol 2002;34:439-58. [Crossref] [PubMed]
- Rhoads JM, Argenzio RA, Chen W, et al. L-glutamine stimulates intestinal cell proliferation and activates mitogen-activated protein kinases. Am J Physiol 1997;272:G943-53. [PubMed]
- Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009;136:521-34. [Crossref] [PubMed]
- Santos AA, Braga-Neto MB, Oliveira MR, et al. Glutamine and alanyl-glutamine increase RhoA expression and reduce Clostridium difficile toxin-A-induced intestinal epithelial cell damage. Biomed Res Int 2013;2013:152052.
- Davidson SM, Papagiannakopoulos T, Olenchock BA, et al. Environment impacts the metabolic dependencies of ras-driven non-small cell lung cancer. Cell Metab 2016;23:517-28. [Crossref] [PubMed]
- Hensley CT, Faubert B, Yuan Q, et al. Metabolic heterogeneity in human lung tumors. Cell 2016;164:681-94. [Crossref] [PubMed]
- Schug ZT, Vande Voorde J, Gottlieb E. The nurture of tumors can drive their metabolic phenotype. Cell Metab 2016;23:391-2. [Crossref] [PubMed]


