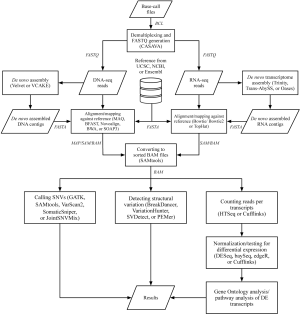
Common applications of next-generation sequencing technologies in genomic research
Introduction
High-throughput molecular analysis is a well-known technology that plays an important role in exploring biological questions in many species, especially in human genomic studies. Over the past 20 years, gene expression profiling, a revolutionary technique, has been widely used for genomic identification, genetic testing, drug discovery, and disease diagnosis, among other things (1). The field of genomics and proteomics research has undergone neoteric fluctuations as a result of next-generation sequencing (NGS), a paradigm-shifting technology that provides higher accuracy, larger throughput and more applications than the microarray platform (2-4). The use of massively parallel sequencing has increasingly been the object of study in recent years. The NGS technologies are implemented for several applications, including whole genome sequencing, de novo assembly sequencing, resequencing, and transcriptome sequencing at the DNA or RNA level. For instance, de novo assembly sequencing assembles the genome of a particular organism without a reference genome sequence (5), which may lead to a better understanding at the genomic level and may assist in predicting genes, protein coding regions, and pathways. In addition, resequencing the organism with a known genome can help in understanding the relationship between genotype and phenotype and identify the differences among reference sequences (6,7). In addition, NGS technologies have been widely used to analyze small RNAs (8-10), including identification of differentially expressed micro RNAs (miRNAs), prediction of novel miRNAs, and annotation of other small non-coding RNAs.
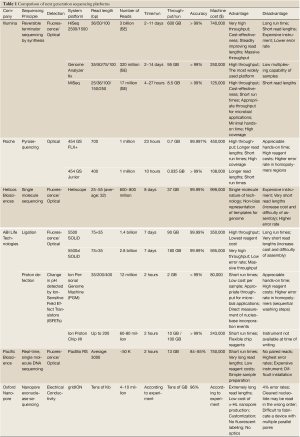
Currently, there are several companies implementing different NGS technologies, such as Illumina (http://www.Illumina.com), Roche (http://www.454.com), ABI Life Technologies (http://www.lifetechnologies.com), Helicos Biosciences (http://www.helicosbio.com), Pacific Bioscience (http://www.pacificbiosciences.com), and Oxford Nanopore (http://www.nanoporetech.com). Table 1 provides a list of some popular NGS instruments and summarizes their respective pros and cons.
Full table
In this review, we begin by discussing preprocessing procedures, i.e., converting raw images into a final set of sequence reads. We then provide an overview of analytic pipelines and recommend some bioinformatics tools that were recently proposed in studies using next-generation DNA and RNA sequencing. Finally, we discuss the small RNA sequencing analytic workflow, annotation databases, and discovery of novel small RNAs by NGS technologies.
DNA sequencing data analysis
Preprocessing procedures
During each run from any NGS platform, several terabytes of raw image data are generated and converted to the FASTQ format files for further analysis. Image analysis uses raw images to locate clusters, export the positions and intensity, and estimate the noise for each cluster. The base-calling step identifies the sequence of base reads from each cluster and filters uncertain or low quality reads. If multiple samples are loaded and run on the same lane, a demultiplexing step is required to identify each sample by its individual index sequences (called “barcodes”). The CASAVA package developed by Illumina handles these preprocessing procedures; likewise, the Bioscope package developed by ABI can be used for preprocessing data in SOLiD format.
Read alignment
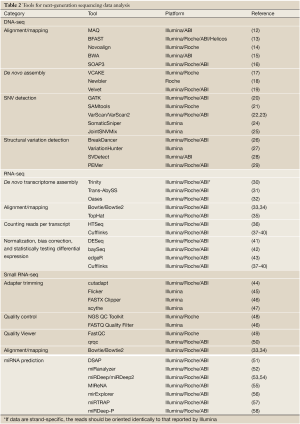
The read alignment in genomics, also called reference-based assembly (11), is utilized by read alignment tools (Table 2) to align several hundred or thousand millions of reads back to an existing reference genome. MAQ (12) is based on the idea of a “spaced seed indexing” strategy to map reads to a reference sequence. BFAST (13) is known for its speed and accuracy on mapping. Novoalign (14) uses the Needleman-Wunsch algorithm and affine gap penalties to find the globally optimum alignment. Burrows-Wheeler Aligner (BWA) (15) is based on Burrows-Wheeler Transformation indexing (59), including the BWA-short algorithm that queries short reads up to ~200 bp with a low error rate and the BWA-SW algorithm that queries long reads with a high error rate. SOAP3, the most recent version of SOAP, supports Graphics Processing Unit (GPU)-based parallel alignment and takes less than 30 seconds for a one-million-read alignment onto the human reference genome (16).
Full table
De novo assembly
The de novo approaches particularly concentrate on grouping short reads into significant contigs and assembling these contigs into scaffolds to reconstruct the original genomic DNA for novel species. The crucial challenge of de novo assembly is that the read length is shorter than repeats in the genome (60). To overcome this problem, three strategies have been proposed (61). First, Warren et al. (17) developed VCAKE, a modification of simple k-mer extension, which is based on the greedy graph approach to assemble millions of reads using high-depth coverage to reduce the error rate. Second, Newbler et al. (18) used the overlap/layout/consensus method to deal with the ambiguous reads within the 454 platform. Lastly, Velvet, a well-known assembler, is applied by the extension of useful graph simplification to reduce the path complexity of the de Bruijn graph (19).
Single nucleotide variant (SNV) detection
After assembling the reads, the next step in analytic pipelines is using a tool to call SNVs for identification of genetic variants. GATK (20) processes re-alignment insertions/deletions (indels), performs base quality recalibration, calls genotypes, and distinguishes true segregating variation by machine learning to discover and genotype variations among multiple samples. SAMtools (21) computes genotype likelihood to call SNVs or short indels. VarScan/VarScan2 (22,23) employs heuristic methods and a statistical test to detect SNVs and indels. SomaticSniper (24) and JointSNVMix (25) use the genotype likelihood model of MAQ and two probabilistic graphical models, respectively, to assess the probability of the differences between tumor and normal genotypes.
Structural variation detection
While SNVs are considered a small genetic change, “structural variation” generally implies a large DNA alteration, approximately 1 kb to 3 Mb in length. Structural variation includes indels, copy-number variants (CNVs), inversions, and translocations (62). A powerful software module for structural variation detection called BreakDancer provides genome-wide screening for large structural variants and detects small indels by integrating evidence across multiple samples and libraries (26). Additional tools are available, such as VariationHunter, which predicts structural variations based on the maximum parsimony principle (27); SVDetect, a chromosomal visualization tool that supports both paired-end and mate-pair sequencing data to predict intra- and inter-chromosomal rearrangements (28); and PEMer, which includes three modules for detection, simulation and annotation of structural variations (29).
RNA sequencing data analysis
Besides exploring the human genome with DNA sequencing (DNA-seq) analysis, high-throughput sequencing has been applied to study RNA transcripts, typically referred to as RNA-seq or transcriptome-seq, and has provided comprehensive knowledge of both genomics and genetics. After the identical preprocessing procedures as in DNA-seq data analysis, RNA-seq data can be used for de novo transcriptome assembly, expression profiling analysis, variant calling and transcriptomic epigenetics.
De novo transcriptome assembly
While de novo DNA assembly is aimed toward building genomic scaffolds for novel species without reference, de novo assembly of RNA-seq data sketches an overview and extracts clues to the “transcriptome.” Current de Bruijn graph-based transcriptome assemblers include Trinity (30), featuring three-step assembly (Inchworm for assembly, Chrysalis for clustering, and Butterfly for processing); Trans-AbySS (31), addressing variation in local read densities; and Oases (32), which introduces dynamic error removal adapted to RNA-seq expression levels. The assembled long RNA contigs can then be annotated with respect to closely related species to fully explore the genome using Basic Local Alignment Search Tool (BLAST), and may serve as a reference for further abundance profiling.
Expression profiling analysis
The predominant application of RNA-seq is currently to profile gene expression levels and identify differentially expressed transcripts among groups of samples. Typical analysis of RNA-seq data for this purpose includes procedures of mapping reads against reference, counting reads per transcript, and statistical testing for differential expression (Figure 1).
In mapping RNA-seq reads, short sequencing reads (FASTQ files) are aligned against the reference sequences (FASTA files), such as annotated genome sequences from the University of California, Santa Cruz (UCSC), the National Center for Biotechnology Information (NCBI), and Ensembl for well-studied species, or against de novo assembled RNA transcripts for novel species. Alignment programs include Bowtie (33), whose ultrafast and memory-efficient method is based on Burrows-Wheeler Transformation indexing (59); Bowtie2 (34), which is improved for finding longer or gapped alignments; and TopHat (35), which adds to Bowtie the capability of integrating known and identifying novel splice junctions. These software packages summarize the aligned results into BAM files, which can be visualized with Integrative Genomics Viewer (63,64).
Taking mapped RNA-seq reads, a Python-based tool, HTSeq (36), extracts read counts for each transcript. The read counts represent raw expression levels of transcripts and are used for statistically testing differential expression among samples subjected to different drug treatments or taken from patients with and without a certain disease. Realizing that the expression distribution of RNA-seq data is different from conventional microarrays (65,66), statisticians and biologists have developed tools for normalizing, bias correcting, and statistically testing RNA-seq read counts of transcripts based on Poisson or negative binomial (NB) distributions. DESeq (41), an R/Bioconductor package based on the NB distribution with adjustments by local regression; baySeq (42), which employs NB statistics and empirical Bayesian approaches; and edgeR (43), which uses the over-dispersed Poisson model combined with empirical Bayesian methods; are three frequently used tools for detecting differential expression of transcripts among a set of sequencing samples. In addition to the mentioned tools, the Cufflinks package (37-40) provides integrated solutions for assembling transcripts, estimating the abundances of transcripts, and testing differential expression. For further biological insights, the differential expression of transcripts can be analyzed for Gene Ontology and pathway enrichment, with methods identical to those implemented in conventional microarray data analyses.
Variant calling and transcriptomic epigenetics
As an alternative to whole-genome DNA sequencing for calling variants (i.e., mutations and single nucleotide polymorphisms), RNA sequencing provides a cost-efficient way for discovering coding variants. Several studies have successfully identified variants in vertebrates from RNA-seq data (67-69). In addition to calling variants upon alignment to reference sequences with SAMtools (21), Hill et al. recently proposed the mutation mapping analysis pipeline for pooled RNA-seq (MMAPPR) (70). With three-step analysis of allele frequency distance calculation, signal processing, and candidate SNP identification, MMAPPR was capable of identifying novel mutants that were biologically validated in zebra fish (70).
Transcriptomic epigenetics via RNA-seq has attracted growing research focus. Efforts in novel research areas, such as transcription start site-associated RNAs (71), promoter-associated RNAs (72), transcription-initiation RNAs (tiRNAs) (73), and long interspersed non-coding RNAs (lincRNAs) (74,75), may facilitate investigations into complex transcriptional regulation (76). However, more bioinformatic and biostatistical input is required before automated tools for complicated RNA-seq data analysis come into practice (77).
Small RNA sequencing
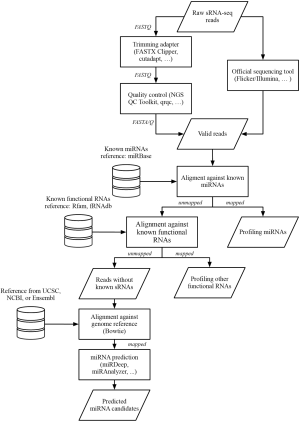
Many classes of small RNAs (sRNAs), such as miRNA, piwi-interacting RNA (piRNA), and small interfering RNA (siRNA), have been reported to play an important role in post-translational regulation of gene expression. Next generation small RNA sequencing (sRNA-seq) technology has now become a gold standard for both sRNA discovery and sRNA profiling, because it is able to sequence the entire complement of sRNAs in a sample with high sensitivity. The following describes a typical workflow and the tools involved.
General workflow
Though the sRNA-seq workflow depends on the application and sequencing platform one uses, some major steps shown in Figure 2 are generally followed. A library consisting of raw cDNA reads is obtained directly after sequencing. First, reads containing sequences of adapters should be trimmed off by using either an official toolkit provided by the company of the sequencer, such as Flicker by Illumina, or third-party toolkits, such as FASTX Clipper of FASTX Toolkit (46), scythe (47), or cutadapt (44). Second, reads having too low overall quality should be discarded using tools like FASTQ Quality Filter of the FASTX Toolkit or the NGS QC Toolkit (48). Next, tools like FastQC (49) or qrqc (50) in R/Bioconductor are used to check the quality statistics visually. Finally, since NGS may produce erroneous reads, the filtered reads should be validated by aligning to a reference genome database. For short read alignment, Bowtie/Bowtie2 are commonly used because they implement an optimized, memory-efficient algorithm and provide many built-in indexes for the genome database reference (33,34).
Some databases are commonly used in sRNA-seq and should be mentioned at the outset. Rfam is an open-access, annotated database providing information about families of non-coding RNAs, such as tRNA, rRNA, and snoRNA (78). miRBase is a database that contains sequences and annotations of all known miRNAs across species; the newest version, miRBase 19, contains around 25,000 mature products in nearly 200 species (79). These two databases help one identify known sRNA reads and one can later choose to either keep these reads or discard them depending on the purpose of the sequencing.
Small RNA prediction
Discovery of new sRNAs is highly facilitated by NGS technology via its massively parallel high throughput of sequencing, which makes it possible to detect sRNAs with lower expression that are hard to find by traditional Sanger sequencing. Methods for identification of miRNAs have been well developed in recent years. There are many distinct algorithms for miRNA prediction, which are implemented in tools such as miRTRAP (57), MIReNA (55), miRExplorer (56), miRAnalyzer (52), miRDeep/miRDeep2 (53,54), and DSAP (51). These tools are mainly designed for animal species. For miRNA prediction in plants, miRDeep-P, a derivative of miRDeep, has been proposed (80). sRNA-seq is also useful in virology (81). Due to the high mutation rates of viruses, sRNA-seq can assist with in silico reconstruction of viral genomes from the antiviral RNAi response and identify virus-derived small interfering RNAs (vsiRNAs) based on the reference sequence (82). Since prediction tools continue to evolve at a fast pace, there is no consensus about which tool is most preferred, and while several comparisons have been made in the aforementioned references, we will defer to the readers to choose the tool most suitable for their situation (83,84).
miRNA characterization
Profiling of miRNAs is another important sRNA-seq application. It has been reported that the miRNA signature can serve as a biomarker for diseases, tissues, or stages of cell development (85,86) and has been used for drug development (58). Currently, microarrays, quantitative real-time RT-PCR, and sRNA-seq are all widely used for miRNA characterization, and their attributes have been described in detail (87). sRNA-seq provides high accuracy for distinguishing miRNAs with similar sequences, such as isomiRs, and can identify novel miRNAs at the same time. However, it should be pointed out that there are reproducible systematic biases toward different protocols of miRNA library construction due to different usage of RNA ligase (88,89). This bias can be eliminated by pooling different adapters (90). Thus, one should be careful about the protocol a dataset uses when performing differential expression analysis across various datasets. Quantitative RT-PCR can be used as a secondary means of absolute quantification.
Future perspectives
NGS technologies provide opportunities for understanding unknown species and complex diseases. Although different companies implement different platforms with distinctive features and advantages, depend on the number of reads and the read length to ensure assembly quality and accuracy. Therefore, an important issue for future research will be the improvement of methods used for analysis of the huge amount of data produced by NGS. The goals will be to increase the accuracy of assembly sequencing, reduce the processing time, and fine-tune the efficiency of algorithms for analysis. In order to make the best use of NGS data, the design of state-of-the-art bioinformatics pipelines to extract meaningful biological insights will be a significant topic in the following years. Ultimately, NGS could reveal human genomic information and help to elucidate the function of the genome, which may provide therapeutic regimens for personalized medicine in the future.
Acknowledgments
We thank Melissa Stauffer, Ph.D., for editing the manuscript.
Funding: We thank National Taiwan University for financial support [grant number: NTU-CESRP-102R7602C7].
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.02.09). EYC serves as the Editor-in-Chief of Translational Cancer Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929-35. [PubMed]
- Git A, Dvinge H, Salmon-Divon M, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010;16:991-1006. [PubMed]
- Roh SW, Abell GC, Kim KH, et al. Comparing microarrays and next-generation sequencing technologies for microbial ecology research. Trends Biotechnol 2010;28:291-9. [PubMed]
- Sîrbu A, Kerr G, Crane M, et al. RNA-Seq vs dual- and single-channel microarray data: sensitivity analysis for differential expression and clustering. PLoS One 2012;7:e50986 [PubMed]
- Li R, Fan W, Tian G, et al. The sequence and de novo assembly of the giant panda genome. Nature 2010;463:311-7. [PubMed]
- Vallender EJ. Expanding whole exome resequencing into non-human primates. Genome Biol 2011;12:R87. [PubMed]
- Voelkerding KV, Dames S, Durtschi JD. Next generation sequencing for clinical diagnostics-principles and application to targeted resequencing for hypertrophic cardiomyopathy: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn 2010;12:539-51. [PubMed]
- Friedländer MR, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 2008;26:407-15. [PubMed]
- Xu G, Wu J, Zhou L, et al. Characterization of the small RNA transcriptomes of androgen dependent and independent prostate cancer cell line by deep sequencing. PLoS One 2010;5:e15519 [PubMed]
- Zywicki M, Bakowska-Zywicka K, Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res 2012;40:4013-24. [PubMed]
- Zhang J, Chiodini R, Badr A, et al. The impact of next-generation sequencing on genomics. J Genet Genomics 2011;38:95-109. [PubMed]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 2008;18:1851-8. [PubMed]
- Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLoS One 2009;4:e7767 [PubMed]
- Novocraft. Novoalign. Available online: http://www.novocraft.com
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [PubMed]
- Liu CM, Wong T, Wu E, et al. SOAP3: ultra-fast GPU-based parallel alignment tool for short reads. Bioinformatics 2012;28:878-9. [PubMed]
- Jeck WR, Reinhardt JA, Baltrus DA, et al. Extending assembly of short DNA sequences to handle error. Bioinformatics 2007;23:2942-4. [PubMed]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005;437:376-80. [PubMed]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008;18:821-9. [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [PubMed]
- Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009;25:2283-5. [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [PubMed]
- Larson DE, Harris CC, Chen K, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 2012;28:311-7. [PubMed]
- Roth A, Ding J, Morin R, et al. JointSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data. Bioinformatics 2012;28:907-13. [PubMed]
- Chen K, Wallis JW, McLellan MD, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 2009;6:677-81. [PubMed]
- Hormozdiari F, Hajirasouliha I, Dao P, et al. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. Bioinformatics 2010;26:i350-7. [PubMed]
- Zeitouni B, Boeva V, Janoueix-Lerosey I, et al. SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics 2010;26:1895-6. [PubMed]
- Korbel JO, Abyzov A, Mu XJ, et al. PEMer: a computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol 2009;10:R23. [PubMed]
- Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 2011;29:644-52. [PubMed]
- Robertson G, Schein J, Chiu R, et al. De novo assembly and analysis of RNA-seq data. Nat Methods 2010;7:909-12. [PubMed]
- Schulz MH, Zerbino DR, Vingron M, et al. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012;28:1086-92. [PubMed]
- Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [PubMed]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357-9. [PubMed]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105-11. [PubMed]
- Anders S. HTSeq: Analysing high-throughput sequencing data with Python. [cited 2013]; Available online: http://www-huber.embl.de/users/anders/HTSeq/
- Roberts A, Trapnell C, Donaghey J, et al. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 2011;12:R22. [PubMed]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511-5. [PubMed]
- Roberts A, Pimentel H, Trapnell C, et al. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 2011;27:2325-9. [PubMed]
- Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46-53. [PubMed]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [PubMed]
- Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 2010;11:422. [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [PubMed]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal 2011;17.
- Illumina. Flicker 3.0. Available online: http://support.illumina.com/sequencing/sequencing_software/flicker_30_small_rna_analysis.ilmn
- Lab H. FASTX-Toolkit. Available online: http://hannonlab.cshl.edu/fastx_toolkit/
- Buffalo V. scythe: A 3'-end Aadapter Contaminant Trimmer. Available online: https://github.com/vsbuffalo/scythe
- Patel RK, Jain M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS One 2012;7:e30619 [PubMed]
- Bioinformatics B. FastQC: A quality control tool for high throughput sequence data.; Available online: http://www.bioinformatics.babraham.ac.uk/
- Buffalo V. qrqc: Quick Read Quality Control. 2012.
- Huang PJ, Liu YC, Lee CC, et al. DSAP: deep-sequencing small RNA analysis pipeline. Nucleic Acids Res 2010;38:W385-91 [PubMed]
- Hackenberg M, Rodríguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res 2011;39:W132-8 [PubMed]
- Friedländer MR, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 2008;26:407-15. [PubMed]
- Friedländer MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 2012;40:37-52. [PubMed]
- Mathelier A, Carbone A. MIReNA: finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics 2010;26:2226-34. [PubMed]
- Guan DG, Liao JY, Qu ZH, et al. mirExplorer: detecting microRNAs from genome and next generation sequencing data using the AdaBoost method with transition probability matrix and combined features. RNA Biol 2011;8:922-34. [PubMed]
- Hendrix D, Levine M, Shi W. miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol 2010;11:R39. [PubMed]
- Eipper-Mains JE, Eipper BA, Mains RE. Global approaches to the role of miRNAs in drug-induced changes in gene expression. Front Genet 2012;3:109. [PubMed]
- Burrows M, Wheeler DJ. A block-sorting lossless data compression algorithm. HP Labs Technical Reports 1994:SRC-RR-124.
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 2012;13:36-46. [PubMed]
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics 2010;95:315-27. [PubMed]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet 2006;7:85-97. [PubMed]
- Thorvaldsdóttir H, Robinson JT, et al. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178-92. [PubMed]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24-6. [PubMed]
- Bullard JH, Purdom E, Hansen KD, et al. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 2010;11:94. [PubMed]
- Marioni JC, Mason CE, Mane SM, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 2008;18:1509-17. [PubMed]
- Miller AC, Obholzer ND, Shah AN, et al. RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res 2013;23:679-86. [PubMed]
- Cánovas A, Rincon G, Islas-Trejo A, et al. SNP discovery in the bovine milk transcriptome using RNA-Seq technology. Mamm Genome 2010;21:592-8. [PubMed]
- Chepelev I, Wei G, Tang Q, et al. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res 2009;37:e106 [PubMed]
- Hill JT, Demarest BL, Bisgrove BW, et al. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res 2013;23:687-97. [PubMed]
- Seila AC, Calabrese JM, Levine SS, et al. Divergent transcription from active promoters. Science 2008;322:1849-51. [PubMed]
- Affymetrix ENCODE Transcriptome Project, Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature 2009;457:1028-32. [PubMed]
- Taft RJ, Glazov EA, Cloonan N, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet 2009;41:572-8. [PubMed]
- Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011;477:295-300. [PubMed]
- Ng JH, Ng HH. LincRNAs join the pluripotency alliance. Nat Genet 2010;42:1035-6. [PubMed]
- Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 2009;10:833-44. [PubMed]
- McGettigan PA. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol 2013;17:4-11. [PubMed]
- Burge SW, Daub J, Eberhardt R, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 2013;41:D226-32. [PubMed]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011;39:D152-7. [PubMed]
- Yang X, Li L. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 2011;27:2614-5. [PubMed]
- Barzon L, Lavezzo E, Militello V, et al. Applications of Next-Generation Sequencing Technologies to Diagnostic Virology. Int J Mol Sci 2011;12:7861-84. [PubMed]
- Vodovar N, Goic B, Blanc H, et al. In silico reconstruction of viral genomes from small RNAs improves virus-derived small interfering RNA profiling. J Virol 2011;85:11016-21. [PubMed]
- Li Y, Zhang Z, Liu F, et al. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res 2012;40:4298-305. [PubMed]
- Williamson V, Kim A, Xie B, et al. Detecting miRNAs in deep-sequencing data: a software performance comparison and evaluation. Brief Bioinform 2013;14:36-45. [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [PubMed]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358-69. [PubMed]
- Git A, Dvinge H, Salmon-Divon M, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010;16:991-1006. [PubMed]
- Linsen SEV, de Wit E, Janssens G, et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 2009;6:474-6. [PubMed]
- Tian G, Yin X, Luo H, et al. Sequencing bias: comparison of different protocols of microRNA library construction. BMC Biotechnol 2010;10:64. [PubMed]
- Jayaprakash AD, Jabado O, Brown BD, et al. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res 2011;39:e141 [PubMed]