
DNA damage tolerance: a double-edged sword guarding the genome
Introduction
DNA, the blueprint of life, encodes the genetic information required for the development, functioning and survival of all known living organisms and many viruses. Damage to DNA impairs essential DNA metabolic processes such as DNA replication, transcription, and recombination and if left unrepaired, leads to events generating chromosomal rearrangements, fusion, deletion, mutations or chromosomal loss. Defective repair or replication errors leads to permanent changes in the genetic information that may pass on to the daughter cells (1). Therefore, a fundamental aspect of life lies in maintaining the integrity of the genome, ensuring the correct inheritance of the genetic information. On one hand, chromosomal aberrations and mutations lead to loss of tumor-suppressors, cell cycle checkpoint genes or result in improper activation of oncogenes, which collectively result in uncontrolled cellular proliferation and development of cancer. However, on the other hand, unrepaired or mis-repaired gross chromosomal aberrations could also result in cellular senescence and cell death, which prevent cancer development (2-4).
DNA damage, ranging from modification of bases, intra- and inter-strand crosslinks, DNA-protein crosslinks, pyrimidine dimers, 6-4 photoproducts, single- and double-stranded breaks, can be caused either by spontaneous changes to the chemical structure of the DNA or by endogenous and exogenous DNA damaging agents (5-9). To counteract the damage inflicted on the genome the cell puts into work a well co-ordinated network of signaling cascade, termed the DNA damage response (DDR). DDR senses the DNA lesion and transmits the damage signal to activate cell cycle checkpoint control in order to delay cell cycle progression and allow for the damage to be repaired by recruiting the DNA repair machinery. Several DNA repair pathways namely, nucleotide excision repair (NER), base excision repair (BER), non-homologous end joining (NHEJ), homologous recombination (HR), and fanconi anemia (FA) exist in the cell that repair different types of DNA lesions (Figure 1). The DNA repair pathways either work independently or co-ordinate to repair complex DNA lesions [reviewed in (10-14)]. Once the damage is repaired the cell re-enters the cell cycle and the genome is duplicated in an error-free manner by high fidelity DNA polymerases and transmitted to daughter cells. During S-phase, as the DNA is being replicated, if the DNA polymerases encounters DNA adducts and/or distortions in the secondary structure of the DNA, the DNA polymerases stalls. Prolonged stalling of replication forks leads to fork collapse and ultimately genome instability. Thus, a second response to damaged DNA, termed DNA damage tolerance (DDT), has evolved to promote replication through and beyond an altered template, leaving the damage to be repaired at a later time point, thus lowering the overall risk of replication fork collapse and genome instability (15,16). In this review, we will describe the DDR signaling cascade that mediates cell cycle checkpoint activation and recruitment of DNA repair factors to sites of DNA damage, especially at stalled replication forks. We will also discuss lesion bypass by DDT pathways and how they act to maintain genome stability. Further, we will describe the pathological consequences that result from a defective response to DNA damage in humans and how different DNA repair pathways can be targeted for cancer therapy.
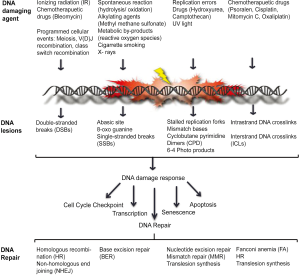
DNA damage response network
DDR is initiated by sensing of DNA damage by DNA damage sensor proteins which then transduce this information to activate cellular responses that includes cell cycle checkpoint control, transcription, activation of DNA repair pathways, senescence and/or apoptosis. Central to this signal transduction cascade are members of phosphatidylinositol 3-kinase-like (PIKKs) protein kinases, ATM, ATR, DNA-PKcs and poly (ADP-ribose) polymerase (PARP) family members PARP1 and PARP2, which respond to distinct DNA lesions and transduce damage signals by modulating many downstream events. In response to DNA DSBs, MRE11-RAD50-NBS1 (MRN) complex functions as the sensor, which recruits and activates, ATM kinase, which in turn phosphorylates effector molecules, such as CHK2, SMC1, H2AX and MRN complex itself to mediate cell cycle checkpoint activation and DNA repair (17-21). DSBs are predominantly repaired by either NHEJ or HR repair pathways. While, NHEJ is an error-prone repair pathway that is mediated by the direct joining of the two broken ends, DSB repair by HR is considered error-free, as it involves processing the DNA ends to generate a 3'-ssDNA overhang that then initiates a homology search for genetic exchange with either the sister or homologous chromosome. Other alternative pathways for DSB repair include alternative NHEJ (alt-NHEJ/MMEJ) and single-strand annealing (SSA) pathways [reviewed in (1,10)].
SSBs generated by IR, reactive oxygen species or indirectly during BER of abasic sites or altered DNA bases, such as 8-oxoG and 3-meA, are sensed by PARP family members (22,23). PARP1 and PARP2 via its zinc finger motifs recognize SSBs and DSBs. Upon binding to DNA, PARP1 and PARP2 get activated and synthesize poly (ADP-ribose) (PAR) chains at sites of DNA damage and on target proteins such as histones H1 and H2B, and PARP1 itself . Histone PARylation contributes to chromatin reorganization and PAR structures in turn serve to recruit factors to promote DNA repair [reviewed in (24,25)].
Signaling stalled replication forks
The most dangerous and frequently occurring lesions in cells are those that arise and result in replication fork stalling during normal DNA replication. During DNA replication, replication forks may stall when they encounter secondary DNA structures, repetitive sequences, certain protein-DNA complexes, or lesions generated by DNA damaging agents. Especially in response to Ultraviolet (UV) induced DNA lesions, replicative DNA polymerases stall since they are unable to accommodate altered DNA bases in their active sites, hindering the progression of DNA polymerases. This can result in the stalling and uncoupling of replicative polymerases with helicase activities of the replication machinery at the lesion site, resulting in tracts of ssDNA due to continuous unwinding of DNA by helicases (26,27). The ssDNA tracts generated are recognized and coated by ssDNA binding protein, Replication protein A (RPA) (27). The direct interaction of ATRIP with RPA coated onto the ssDNA serves to recruit ATR-ATRIP kinase complex at DNA damage sites (28). Work from several groups has helped understand the intricate cross-talk between signaling components at stalled replication forks (Figure 2). Following recruitment, ATR is partially activated by autophosphorylation of ATR at Thr-1989, a process dependent on RPA, ATRIP and ATR kinase activity (29). The full activation of ATR-ATRIP requires additional mediators that include RAD17, the RAD9-RAD1-HUS1 [9-1-1] checkpoint clamp, TopBP1 and BACH1 at the damage site (30-33). The initial recruitment of TopBP1 to sites of stalled forks is signaled by RPA-ssDNA. However, in a recent finding it was shown that γ-H2AX plays an indirect role in the recruitment of TopBP1 in response to replication stress. Upon DNA damage H2AX accumulates on dsDNA region adjoining ssDNA at stalled forks and is phosphorylated by ATR or other related kinases, which triggers the recruitment of MDC1. MDC1 then leads to the accumulation of TopBP1 at stalled replication forks via direct protein-protein interaction (34,35). TopBP1 interacts with p-BACH1 (Thr-13) at the damage sites. BACH1 is recruited to the sites of damage, independent of TopBP1. It is proposed that BACH1 together with TopBP1 may facilitate the unwinding of dsDNA or other DNA structures at stalled forks, generating long tracts of ssDNA which is subsequently coated by additional RPA molecules (36). This accumulation of RPA-ssDNA is required for the amplification of replication stress signals leading to the recruitment of ATRIP-ATR and the 9-1-1 complex (36,37). Independent of ATR, Rad17-RFC complex recognizes ssDNA-dsDNA junctions and loads 9-1-1 complex onto dsDNA (38,39). RAD9 in the 9-1-1 complex interacts with and further recruits TopBP1 to the damage sites (40,41). TopBP1 and 9-1-1 complex localized on dsDNA, now communicate with ATR-ATRIP recruited on ssDNA of the damaged fork via TopBP1. TopBP1 stably interacts with phosphorylated ATR via the BRCT domains 7 and 8 and thereby fully activates the ATR-ATRIP complex at damage sites. Additionally, phospho-ATR tethers TopBP1 to stimulate multiple ATR-ATRIP complexes on RPA-ssDNA (29,42). Active ATR kinase can then phosphorylate several components of the replisome, RPA, Claspin (43-45) and also it’s downstream effector kinase, CHK1 (46,47). As well demonstrated in budding yeast, full activation of the ATR-Chk1 (mec1/rad53) checkpoint signaling pathway is also mediated by Tim1 (Tof1), Tipin (Csm3) and Claspin (Mrc1) (48-54). Both Timeless and Tipin are required for normal replication and ATR-Chk1 activation in response to replication stress (52,55,56). Tipin binds to RPA2 subunit of the RPA complex (RPA1, RPA2 and RPA3) and this interaction is required for the stable association of Timeless-Tipin and Tipin-Claspin complexes on RPA-coated ssDNA (57). In response to replication stress, Claspin localized on RPA coated ssDNA is phosphorylated by ATR and this phosphorylation is required for its interaction with Chk1, thereby serving as a platform for ATR mediated phosphorylation and activation of Chk1 upon DNA damage (58-60). ATR mediated phosphorylation of the chromatin bound Chk1 at S/T-Q sites (S317, S345 and S366), is required to stimulate the kinase activity of Chk1 and its subsequent dissociation from the chromatin, facilitating the transmission of the DNA damage signal to downstream targets, thereby promoting efficient cell cycle arrest in response to DNA damage (61-63) (Figure 2). Checkpoint kinases, Chk1 and Chk2, initiate cell cycle arrest primarily by inhibiting Cyclin-Cdk complex activity (Cdk4/cyclin D and Cdk2/cyclin E for G1-phase checkpoint, Cdk2/cyclin A for S-phase checkpoint, and Cdc2/cyclin A and Cdc2/cyclin B for G2-phase checkpoint) upon DNA damage until the DNA damage is repaired [reviewed in (64-66)].
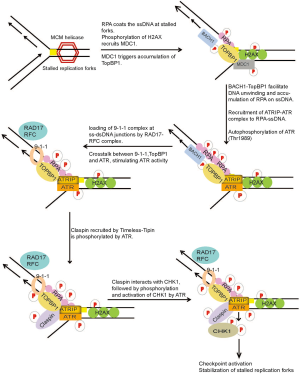
Replication fork stalling can directly generate ssDNA; however, ssDNA structure can also arise during NER, repair of ICLs and at dysfunctional telomeres. In addition, ATR-Chk1 pathway is also activated in response to DSBs, when ssDNA is generated as a result of resection of DSB ends by exonucleases during DNA repair by HR pathway. Further, replication of damaged DNA can result in DSBs when leading-strand DNA polymerases encounter single-strand nicks or abasic sites (27). Thus, although the ATM-Chk2 and ATR-Chk1 pathways are frequently activated together in cells exposed to diverse genotoxic stresses, genetic and biochemical studies show that ATR-Chk1 is the principal effector involved in diverse DNA damage and replication checkpoint controls, while ATM-Chk2 function is mainly restricted to DSBs (67,68). Therefore, it is not surprising that germ-line inactivation of ATR or Chk1, but not ATM or Chk2, result in early embryonic lethality (69-72). Similarly, depletion or inactivation of ATR or Chk1, but not ATM or Chk2, affects cell viability (47,71-73). Interestingly, DT40 lymphoma cells survive genetic inactivation of Chk1, albeit with impaired cell growth and survival (73), suggesting that cells may find ways to bypass this requirement, which may have implications for the development of resistance when CHK1 inhibitors enter in clinical trials.
Stabilization of stalled replication forks and replication restart
Stalled forks are fragile structures and thus prolonged stalling of replication forks can lead to fork collapse, generation of DSBs and genome instability. Therefore, stabilization and rescue of stalled forks and subsequent completion of DNA replication is essential for cell survival and genome maintenance. RPA coats the ssDNA generated at the stalled forks and signals the recruitment of proteins that stabilize and restart stalled replication forks. Recently, several groups showed that SMARCAL1 and AH2, a new class of enzymes that catalyze the rewinding of RPA-coated ssDNA, are required to stabilize stalled forks in response to replication stress (74-82). While, SMARCAL1 is recruited to damage sites by RPA, AH2 recruitment is mediated by proliferating cell nuclear antigen (PCNA) (74-78,80-82). The annealing activity of SMARCAL1 and AH2 was shown to be required for stabilizing stalled replication forks. Restart of stalled forks is proposed to require the action of several helicase and translocases that include BLM, WRN, FANCM and HLTF that are also recruited by RPA (83-85). BLM, WRN, FANCM, and HLTF have been suggested to regress replication forks by favoring the annealing of the leading- and lagging strands and generating a HJ-like structure also known as ‘‘chicken foot,’’ which could allow the restart of DNA synthesis by template switching and lesion bypass (86,87). SMARCAL1 might also promote replication fork regression, given the similarity of its helicase domain with that of yeast Rad5, which is known to regress replication forks (86,87). However, future studies are needed to reveal the mechanism by which these proteins coordinate and prevent fork collapse at stalled replication forks.
DNA damage tolerance pathway
Restart of stalled replication forks is achieved by DNA damage tolerance pathways, also known as post replication repair (PRR) that initiate DNA synthesis downstream of the DNA lesion and therefore allow the lesion to be repaired after DNA replication. Earlier studies in both yeast and mammalian cells suggest two major pathways for PRR: translesion synthesis (TLS) and the damage avoidance by template switching (TS) (15,88-90) (Figure 3). The replicative DNA polymerases are particularly specific for normal DNA base pairs, but cannot accommodate damage bases or bulky adducts into their active site. During TLS, the stalled replicative polymerase is replaced by TLS polymerases, which are a class of specialized polymerases with low-processivity but can replicate over distortions in DNA and directly bypass lesions. Mammalian cells have at least seven enzymes with TLS activity. These include four Y-family polymerases Pol η (POLH), Pol ι (POLI), Pol κ (POLK) and REV1, one B-family polymerase, Pol ζ, the catalytic subunit of which is REV3L and two A-family polymerases Pol θ (POLQ) and Pol ν (POLN). Each of the TLS polymerases has different substrate specificities for different types of DNA damage (16,91,92). For example, Pol η preferentially inserts two adenines opposite a thymine dimer, while Pol κ has been shown to accurately bypass benzopyrene-induced guanine adducts and Pol ζ extends distorted base pairs, such as mismatches that might result from an inaccurate base insertion by a TLS polymerase or a base pair involving a bulky DNA lesion (93-96). Therefore, depending on the TLS polymerase that is recruited, lesions can be bypassed either in a relatively error-free mode, for example when using DNA polymerase eta (Pol η), or by an error-prone mechanism using Pol ζ and Rev1 (96,97). The mechanism of lesion bypass by damage avoidance is unclear, but is thought to involve template switching with the undamaged sister chromatid and/or the use of HR pathway (15,85). Nevertheless, both of these direct (TLS) and indirect (TS) bypass pathways allow for resumption of DNA replication and leave lesions for repair at a later time point (Figure 3).
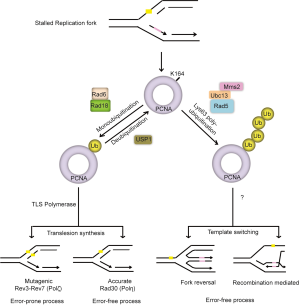
PCNA ubiquitination
A critical step in the regulation of PRR is the posttranslational modification of PCNA, the replicative sliding clamp that plays an essential role in DNA replication. Following DNA damage and/or replication stress, PCNA is either mono- or poly- ubiquitinated on lysine 164 (K164) (89,98-100). Studies suggest that monoubiquitination of PCNA promotes direct lesion bypass by recruiting TLS polymerases to stalled replication forks (97,101-103), while polyubiquitination of PCNA promotes damage avoidance through a process that is still unclear (98,104) (Figure 3). In yeast, ubiquitination of PCNA is mediated by the Rad6 epistasis group and two RING domain-containing E3 ligases, Rad18 and Rad5. Rad18 mediates the monoubiquitination of PCNA, while Rad5 facilitates the further addition of K63-linked ub-chains (97,101,102,105,106). In humans, mono-ubiquitination on lys-164 is the major modification of PCNA detected, which is carried out by RAD18 upon exposure of replicating cells to DNA damage induced by UV or hydroxyurea (HU) (107). Polyubiquitination of PCNA detected at much lower levels is believed to be mediated by HLTF and SHPRH, the mammalian orthologs of yeast Rad5 ubiquitin ligase (85,108). Recent finding challenges this view and suggests that HTLF, which transfers polyubiquitin chains to RAD6, permits RAD18 to attach polyubiquitin K63-linked ubiquitin chains to PCNA. Thus, RAD18 may directly control both states of PCNA ubiquitination (109). However, the exact mechanism underlying ubiquitin chain elongation on PCNA and what triggers the switch from mono- to poly-ubiquitination of PCNA remains to be elucidated. Furthermore, how polyubiquitination of PCNA via K63-linked serves as a signal to recruit factors involved in error-free PRR and factors that activate and regulate translesion synthesis and template switching processes remains to be determined.
RPA-coated ssDNA induces ubiquitination of PCNA. However, it is not clear how checkpoint and DNA damage tolerance processes are coordinated at stalled replication forks (110,111). As discussed in the early section, RPA-ssDNA is a central component for ATR-CHK1 checkpoint pathway in response to replication stress. Interestingly, studies show that claspin and Chk1, but not ATR, is required for PCNA ubiquitination (112,113). Thus, it is not yet clear whether the checkpoint proteins directly or indirectly affect the recruitment of RAD18 to RPA-ssDNA. It could be possible that RPA-ssDNA initiates two DNA damage response pathways in parallel: the ATR-dependent checkpoint control and PCNA-mediated DNA damage tolerance, wherein each pathway assembles its own RPA-ssDNA-clamp: RPA-ssDNA-RAD18 and PCNA for PCNA ubiquitination, and the RPA-ssDNA-ATRIP and 9-1-1 checkpoint clamp for ATR activation (114,115). A single stalled fork may be able to accommodate both complexes simultaneously, as 9-1-1 complex loads preferentially onto the 5' end of the primed RPA-ssDNA during checkpoint activation, while PCNA occupies the 3' end during processive DNA replication. Thus, at least part of the solution may lie in clamp specificity (116,117). Moreover, although ATR activity appears to be dispensable for PCNA ubiquitination, genetic studies in yeast suggest a role for the ATR checkpoint pathway in regulating TLS polymerase activity independent of PCNA monoubiquitination (118,119). Thus, the connections between the checkpoint and damage tolerance pathways; especially regarding their recruitment, activity and regulation, are largely unexplored and needs further investigation.
Translesion synthesis
Monoubiquitination of PCNA (ub-PCNA) increases its affinity for TLS polymerases Polη, Polι, and Rev1 (101,107,120,121). This increase of affinity for ub-PCNA is mediated by ubiquitin-binding domains (UBM or UBZ domains) identified in all of the Y-family polymerases and therefore provide a possible mechanism for polymerase switch, whereby the blocked replicative DNA polymerase is replaced by a TLS polymerase that can bypass the lesion (101,107,122-124). Thus, the current model for TLS process involves at least two polymerase-switching events, in a step-wise fashion. In the first switch, the stalled replicative DNA polymerase is replaced by a TLS polymerase capable of replicating over the DNA lesion. The TLS ‘patch’ is then extended by either the same or another TLS polymerase. The extension step allows for the lesion to escape detection by the 3' to 5' exonuclease proofreading activity of the replicative DNA polymerase. This is followed by the second switch, which restores the high fidelity replicative DNA polymerase to the DNA template to resume DNA replication (15,16,96,125) (Figure 4).
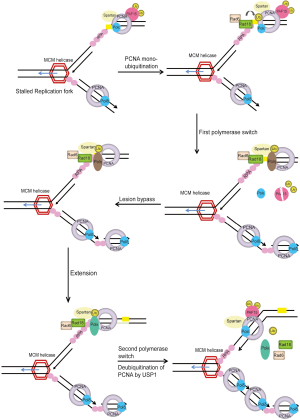
PCNA ubiquitination is the key event regulating PRR, however, it is insufficient, by itself, to account for the specificity of PRR pathway choice, as several TLS polymerases have ubiquitin-interacting motifs (97). Recently, finding from several groups identified Spartan (C1orf124/DVC1) as a regulator of TLS (126-132). Spartan co-localizes and interacts with unmodified and ub-PCNA at damage sites, which require the PIP box and UBZ domain of Spartan. Spartan was shown to stabilize RAD18 and ub-PCNA at sites of DNA damage (126-132). Interestingly, Spartan binds to replicative DNA polymerase POLD3 and PDIP1 under normal conditions but preferentially associates with TLS polymerase η (POLH) upon UV damage (128). Depletion of Spartan induced the association of POLD3 with Rev1 and the error-prone TLS polymerase Pol ζ, and led to elevated mutagenesis that requires POLD3, Rev1 and Pol ζ (132). Spartan also binds to the AAA-ATPase VCP (p97) via its SHP domain and recruits p97 to sites of DNA damage where p97 may facilitate the extraction of the TLS polymerase Polη during DNA repair to prevent excessive TLS activity and therefore limit the incidence of mutations caused by the action of TLS polymerases (126-132). Thus, Spartan plays important roles at several steps in TLS, it is required to stabilize RAD18 and PCNA at damage sites; it promotes the switch from replicative to translesion polymerase; and it is required to prevent excessive TLS activity and suppress mutations caused by lesion bypass (Figure 4) (126-132).
Another regulator of TLS identified recently is PCNA-associated factor PAF15 (KIAA0101) (133). Study showed that in unperturbed S phase, PAF15 is modified by double mono-ubiquitylation of Lys-15 and 24 sites and is tightly bound to PCNA on the chromatin. Replication stress triggers rapid, proteasome-dependent removal of lys 15/24-ubiquitylated PAF15 from PCNA, facilitating lesion bypass by allowing the recruitment of TLS polymerase Polη to ub-PCNA at stalled replisomes (Figure 4) (133). Monoubiquitinated PCNA is shown to be deubiquitinated by the cysteine protease Usp1 (134). Thus, TLS may be regulated by both ubiquitination and deubiquitination. Interestingly, Usp1 degradation is observed after UV treatment but not after treatment with methylmethanesulfonate (MMS) or mitomycin C (MMC), which suggests that deubiquitination by Usp1 may be a UV-specific mode of regulating TLS and that other possible deubiquitinases might function in response to different types of genotoxic stress (Figure 4) (113).
Thus, with several new regulators identified to function in TLS synthesis, several important questions come into sight. Are there more TLS regulators in mammalian cells? Do these regulators talk to each other? How are the functions of TLS regulators such as Spartan and PAF15 regulated and how do they act with repair factors? These are all important biological questions that remain to be addressed. Moreover, while the molecular and physiological roles of some of the TLS polymerases such as Polη, REV1 and REV3L have been studied, the functions of POLN, POLK, POLI and POLQ in lesion bypass still need to be established.
TLS polymerases in ICL repair and genome maintenance
Studies in chicken DT40 and mammalian cells have shown that REV7, REV3 and REV1 are required for cellular survival to genotoxic stress such as UV, MMS, DNA crosslinking agents, and DSBs (135-141). These suggest that Rev1 and Pol ζ have additional roles in promoting survival beyond the conical PRR pathway.
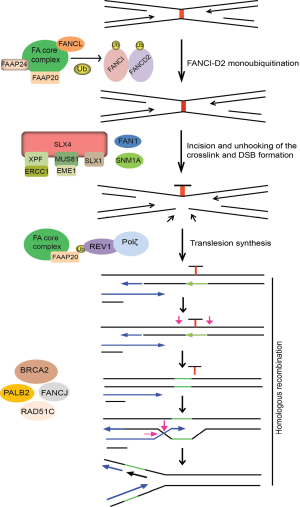
The current model for ICL repair involves the cooperation between the FA pathway, translesion DNA synthesis by Rev1 and Pol ζ, and HR. Central to ICL repair is FA pathway that comprises of 15 FA genes (FANC-A, -B, -C, -D1/BRCA2, -D2, -E, -F, -G, -I, -J/BRIP1/BACH1, -L, -M, -N/PALB2, -O/RAD51C, and -P) and five Fanconi Anemia Associated Proteins (FAAP-20, -24, -100, MHF1 and MHF2) (13,142). The critical step in FA pathway is the monoubiquitination and subsequent activation of the FANCI and FANCD2 (I-D2) complex at the ICL site, which is mediated by the FA core complex, comprising of eight FA proteins (FANC-A, B, C, E, F, G, L and M) and FAAP20, -24 and FAAP100, with FANCL serving as the E3 ligase (13,143). Following ubiquitination, the I-D2 complex is recruited to the chromatin and signals the recruitment of FAN1 nuclease to ICL site. FAN1 is recruited to ICL site via its interaction with ub-ID2 through its ubiquitin binding ZNF domain. FAN1 displays 5'-flap endonuclease and 5'-3' exonuclease activity and once targeted to the chromatin, FAN1 may act alone or together with other structure-specific nucleases (SSEs) to mediate endonucleolytic digestion of cross-linked DNA (144-147).
Incision around the ICL is a key step in initiating the repair process. A number of SSEs have been implicated in the incision events during ICL repair, which include XPF-ERCC1, MUS81-EME1 and hSNM1A (148-152). SLX4 (FANCP) serves as a scaffold protein for three SSEs, XPF-ERCC1, MUS81-EME1 and SLX1 (153-156). The exonuclease SNM1A is biochemically active at sites of crosslinks and required for ICL resistance, which is consistent with an in vivo function of this nuclease in ICL repair. However, whether and how SNM1A interacts and acts with FA proteins remains largely unknown, although it is speculated to act in concert with SLX4-associated XPF-ERCC1 to process the crosslink after the initial incision (157). Thus, several SSEs are believed to function in incision and unhooking of ICLs. But, whether these nucleases act in parallel or in sequence in this process and how they are recruited and regulated during ICL repair have not been fully elucidated. The downstream events following ICL incision and unhooking are believed to be mediated by BRCA2, FANCJ, PALB2 and RAD51C, which essentially function in HR.
Direct evidence implicating the involvement of Pol ζ in ICL repair was obtained using the Xenopus egg extract system, where it was shown that ubFANCI-D2 promotes both the incision and TLS steps of ICL repair, suggesting that like PCNA ubiquitination, FANCI-D2 ubiquitination may play a role in recruiting TLS polymerases to ICLs via their ubiquitin-binding domains (158,159). However to date, no evidence exists suggesting a direct interaction between FANCD2 and Rev1. Nevertheless, cells from FA patients are hypomutagenic in response to UV, similar to Rev1 and Rev3-deficient cells (160). Also, FANCC-deleted DT40 cells have a lower frequency of spontaneous point mutations, suggesting that an intact FA pathway may be required for Rev1-mediated error prone TLS (161). Overall, these results imply that Rev1 and Rev3 are regulated by FA pathway to execute lesion bypass in two different scenarios: bypass of UV-generated photoproducts or cisplatin induced intra-strand crosslinks and the repair of ICLs. However, there are differences in the requirement of FA proteins for TLS in response to replication blocks versus ICL repair. The recruitment of Rev1 to damage sites following UV and cisplatin treatment requires an intact FA core complex but is independent of FANCD2 monoubiquitination, suggesting that FA core complex alone regulates TLS in response to replication fork stalling by UV or cisplatin (136,162). Accordingly, patient cells deficient in FA core components generate spontaneous and UV-induced point mutations at a low frequency compared to cDNA corrected cells (162). Recent findings identified FAAP20 to be an integral part of the FA core complex which plays a role in maintaining the integrity of the core complex, and promotes FANCD2 monoubiquitination (163,164). Studies showed that FAAP20 directly binds to FANCA and the UBZ domain of FAAP20 associates with ub-Rev1, providing a physical link between Rev1 and the FA pathways (163). Together, these studies suggest that FAAP20 specifically promotes Rev1-dependent TLS across replication stalling lesions like thymine dimers and bulky cisplatin adducts and may also directly promote Rev1-dependent TLS during ICL repair (Figure 5). Further, depletion of REV1, REV3 or REV7 in human cells is associated with a reduction in HR efficiency by approximately 50%, similar to FA cells, as opposed to 90-95% seen in cells deficient in RAD51 protein (137). This similar degree of reduction in HR repair is observed in cells depleted of another DNA polymerase POLN that is implicated in ICL repair and HR. Moreover, no additive reduction in gene conversion efficiency is observed upon FANCD2-POLN co-depletion, suggesting that POLN participates in the same FA pathway that regulates, but not essential for, HR repair (165). Since these studies measure repair of a site specific DSB by HR, they imply that alternative DNA polymerases may be needed to perform a subset of these reactions and are not confined to preparing the sister chromatid for HR repair after ICL unhooking. Together, these observations suggest that Rev1 and Pol ζ (possibly in collaboration with POLN) may be required to synthesize DNA during a subset of HR reactions that involve extension of distorted or mis-paired primer templates that would otherwise cause stalling of normal DNA polymerases. At least in mammalian cells, the FA pathway may be important for regulating TLS during HR repair and rescues problematic HR templates by recruiting the TLS pathway.
DDR defects and cancer
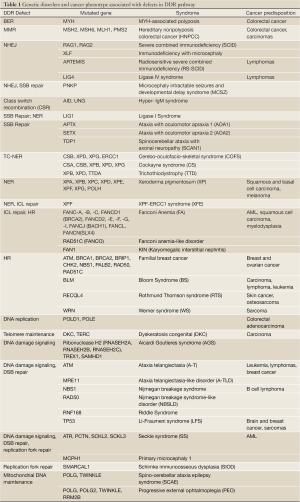
The importance of DDR in human physiology is highlighted by the broad spectrum of human diseases caused by mutations in DDR genes. The phenotypes caused by DDR defects range from neurological dysfunction, immunodeficiency, reproductive and growth defects, premature ageing and cancer (166-171) (Table 1). Since genomic instability is one of the key drivers leading to cancer development (172,173), it is not surprising that most genetic disorders resulting from defective mutation in one or more DDR genes predispose patients to cancer development (174). Somatic mutations in DDR genes are observed in several types of cancer. Hereditary nonpolyposis colorectal cancer (HNPCC) is caused by heterozygous mutations of MMR genes, such as MLH1, MSH2, MSH6, and in fewer cases, PMS2 (175). The most prevalent mutations leading to hereditary breast and ovarian cancer affect the HR genes BRCA1 and BRCA2 (176-178). A broad spectrum of malignancies is displayed by the DDR disorders like Bloom syndrome (BLM), Werner syndrome (WRN), Rothmund Thomson syndrome (RECQL4) and Li-Fraumeni syndrome (TP53) (179,180).
Full table
The Y family polymerases are responsible for the majority of mutagenic events and hence play a central part in carcinogenesis. As overactive TLS leads to increased mutagenesis, it can drive a normal cell to become cancerous and also can cause chemo-resistant in cancer cells. On the other hand, reduced TLS activity can lead to increased fork stalling and DSB formation in the genome, triggering genome instability and cancer development. Thus, DNA damage tolerance pathway functions as a double-edged sword guarding the genome. TLS polymerase POLH is directly linked to tumor suppression in humans. XPV, a rare inherited human disorder characterized by increased incidence of sunlight-induced skin cancers is caused by inactivating mutations of POLH (181). In the absence of Pol η activity, XPV cells cannot perform accurate bypass of ultraviolet UV induced TT-dimers, and the resulting mutations at TT sites are thought to cause skin cancer (103,182,183). Further, expression of POLH is found to be greatly reduced in lung and stomach cancers, while increased POLH expression correlated with poor clinical outcomes (184,185). Increased expression of POLB has now been observed in gastric, uterine, prostate, ovarian and thyroid carcinomas (186-188). Increased POLQ expression in breast cancer tumor samples correlates with poor clinical outcome (189). Apart from TLS polymerases, germ-line mutations affecting the proofreading domain of replicative polymerase POLD1 and POLE also predispose carriers to colorectal adenomas and carcinomas (190).
Cancer therapeutics essentially comprises of surgery in combination with radiotherapy and/or chemotherapy. The mode of action of most chemotherapy drugs relies on the induction of DNA damage in rapidly cycling tumor cells with inadequate DNA repair. For example, platinum drugs, which are particularly active against germ cell tumors and serous ovarian cancer, induce intra- and inter-strand crosslinks in DNA. Anthracyclines induce damage by inhibiting DNA topoisomerases and by producing oxygen radicals. DNA lesions are usually quickly recognized by DDR factors and repaired in normal cells. However, cancer cells often have defective DDR, which leads to genome instability and cancer development. These DDR defects favor cancer therapeutics since they allow preferential killing of tumor cells (191,192). Unfortunately, DDR defects can also lead to acquired drug resistance during treatment. To overcome this problem, increasing effort is being directed to sensitize tumor cells to DNA damaging agents by overriding checkpoint activation, as replication in the presence of DNA damage would lead to mitotic catastrophe and cell death in the absence of DNA damage checkpoints. Potential inhibitors targeting ATM (KU55933, KU60019 and CP466722), ATR (VE-821 and NU6027), CHK1 (UCN-01, G06976), CHK2, WEE1 (MK1775) and CDC25 (IRC-083864) to override cell cycle checkpoints have been developed and are now in clinical trials [reviewed in (192)]. Other strategies, such as pharmacological inhibition of the interaction between p53 and its negative regulator MDM2 by nutlin, RITA or reactivation of mutant p53 by PRIMA1 (193-196) in p53 deficient tumors are also being evaluated. Furthermore, demethylating compounds such as 5-azacytidine can activate the cyclin-dependent kinase 2A (Cdkn2a) locus, which is frequently hypermethylated in many cancers, lead to the repression of INK4A and ARF, and therefore reinforce G1/S cell cycle checkpoint and inhibit tumor proliferation (197).
Several inhibitors that directly block DNA repair pathways have also been identified for cancer therapy. BER pathway repairs DNA damage that is therapeutically induced by IR, DNA methylating agents and topoisomerase I poisons (camptothecin, irinotecan and topotecan). Inhibitors that target various components of BER components include FEN1, ligase 1 and ligase 3 have shown to enhance sensitivity of tumors to IR, TMZ and MMS, respectively (198-200). The most advanced and promising drugs currently targeting DNA repair are AP endonuclease 1 (APE1/APEX1) inhibitors and PARP inhibitors (PARPis) (201-203). Similarly, inhibitors targeting other DNA repair pathways like NHEJ, HR and NER have also been identified and are under study (191,192). Moreover, HDACs are also being tested as druggable targets for inhibiting DNA repair. For example, HDAC inhibitors such as vorinostat may be used as chemosensitizers (192,204,205).
The most promising prospect for future cancer treatment is the exploitation of synthetic lethality that targets specific DDR defects. The best example is the use of PARP inhibitors olaparib, which show additive or synergistic activity in preclinical models of BRCA-deficient cancer (206-209). Knockdown of HR pathway genes such as RAD51, RPA, NBS1 and CHK1 also conferred sensitivity to PARPis, expanding the range of potential targets for PARPi therapy (210). Similarly, inhibition of BER with APE1 inhibitors is also synthetically lethal in cells with HRR dysfunction (211). Other examples of synthetic lethality of HRR defects with PARPis are also emerging such as loss of ATM or MRE11, which occurs secondary to MMR defects in tumors, confers sensitivity to PARPis (212-214). Moreover, ATM knockdown, or inhibition with KU55933, is synthetically lethal in cells with defects in FA pathway. Additionally, CHK1 inhibitor G06976 reduced cell survival and profoundly increased cisplatin sensitivity in cells with defective FA pathway, thus raising the potential for targeted therapy using ATM or CHK1 inhibitors in patients with defective FA pathway (215,216).
Effort is also being directed to target DNA polymerases for chemotherapy. For example, dehydroaltenusin derivatives inhibit Pol α, but not Pol β, Pol δ, Pol ε or Pol γ, and are cytotoxic in tumor cells and in xenograft tumor models (217,218). Eicosapentaenoic acid is an inhibitor of Pol β, Pol δ and Pol ε and radiosensitizes cells (219). Several small-molecule inhibitors of Pol β have been found that can potentiate the toxic effects of the chemotherapeutic drugs bleomycin and temozolomide (199,220,221). Reduction of POLB expression increases sensitivity to the chemotherapeutic agent oxaliplatin (222). The TLS DNA polymerases are also possible targets for enhancing DNA-damaging therapies, and inhibitors of some of these are beginning to emerge (91,223). Suppression of Rev1 and Rev3l expression leads to increased sensitivity to cisplatin and cyclophosphamide in a xenograft model, and tumors in which Rev1 expression is knocked down do not develop resistance to cyclophosphamide (224). Rev3l downregulation increased the sensitivity to cisplatin in a xenograft model of non-small-cell lung cancer (225). Thus, these initial findings indicate that inhibition of the Y family polymerases results in a combination of hyper-sensitivity to killing by DNA damaging agents and, in some cases, reducing damage-induced mutations, suggesting that inhibiting their function may not only sensitize tumors to DNA-damaging chemotherapeutics, but also reduce the incidence of chemotherapy-induced secondary tumors and prevent the acquisition of resistance to chemotherapy. These potentials make Y-family polymerases attractive cancer drug targets. Of course, a thorough analysis will be needed to determine the promise and limitations of inhibiting TLS as a potential approach for cancer treatment.
Conclusions
Work over several decades has brought to light important key players, post-translational modifications, cross-talk and co-ordination of several cellular processes in responding, repairing DNA lesions and preserving the integrity of our genetic information. With more knowledge come more questions. With multiple repair pathways, each functioning alone or in co-ordination with one or many other pathways to repair complex lesions such as ICLs, the degree of complexity increases with every step. How do different repair pathways co-ordinate and how are all these pathways regulated as a whole and at the level of each and every signaling and repair step or process? How chromatin context and cell-cycle phase influence repair outcome? How are the DNA lesions repaired within the context of active DNA replication? These are questions that remain a challenge to the field and will be major focuses for future studies. With DDR network being tested as promising targets for cancer therapy, their cytotoxicity, therapy resistance and generation of secondary cancers are being added to these complexities. The rapid advances in genome sequencing and other cutting-edge technologies may make it possible to identify critical mutations in a given tumor prior to therapy, and therefore enable a more targeted approach for the use of current and expanding arsenal of DNA damaging agents as therapeutic drugs. Understanding the basic biology underlying DDR with the careful and calculated use of DDR targets will provide better and efficient therapy for cancer and other human diseases.
Acknowledgments
We apologize to those colleagues whose work has not been cited due to space limitation.
Funding: This work was supported by grants from the National Institutes of Health (CA089239, CA092312, and CA100109 to J.C.). J.C is also a recipient of an Era of Hope Scholar award from the Department of Defense (W81XWH-05-1-0470) and a member of M.D. Anderson Cancer Center (CA016672).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (David J. Chen and Benjamin P.C. Chen) for the series “DNA Damage and Repair” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.04.01). The series “DNA Damage and Repair” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kasparek TR, Humphrey TC. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol 2011;22:886-97. [PubMed]
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 2007;26:7773-9. [PubMed]
- Moraes MC, Neto JB, Menck CF. DNA repair mechanisms protect our genome from carcinogenesis. Front Biosci (Landmark Ed) 2012;17:1362-88. [PubMed]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med 2009;361:1475-85. [PubMed]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993;362:709-15. [PubMed]
- Jena NR. DNA damage by reactive species: Mechanisms, mutation and repair. J Biosci 2012;37:503-17. [PubMed]
- Joenje H. Metabolism: alcohol, DNA and disease. Nature 2011;475:45-6. [PubMed]
- Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans 2006;34:523-5. [PubMed]
- Lieber MR, Chang CP, Gallo M, et al. The mechanism of V(D)J recombination: site-specificity, reaction fidelity and immunologic diversity. Semin Immunol 1994;6:143-53. [PubMed]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet 2011;45:247-71. [PubMed]
- Dianov GL, Hübscher U. Mammalian Base Excision Repair: the Forgotten Archangel. Nucleic Acids Res 2013;41:3483-90. [PubMed]
- Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genet 2012;28:566-73. [PubMed]
- Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 2013;493:356-63. [PubMed]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010;40:179-204. [PubMed]
- Chang DJ, Cimprich KA. DNA damage tolerance: when it's OK to make mistakes. Nat Chem Biol 2009;5:82-90. [PubMed]
- Sharma S, Helchowski CM, Canman CE. The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability. Mutat Res 2013;743-744:97-110. [PubMed]
- Matsuoka S, Rotman G, Ogawa A, et al. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A 2000;97:10389-94. [PubMed]
- Yazdi PT, Wang Y, Zhao S, et al. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev 2002;16:571-82. [PubMed]
- Uziel T, Lerenthal Y, Moyal L, et al. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 2003;22:5612-21. [PubMed]
- Burma S, Chen BP, Murphy M, et al. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 2001;276:42462-7. [PubMed]
- Smith J, Tho LM, Xu N, et al. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 2010;108:73-112. [PubMed]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet 2008;9:619-31. [PubMed]
- Miwa M, Masutani M. PolyADP-ribosylation and cancer. Cancer Sci 2007;98:1528-35. [PubMed]
- Sousa FG, Matuo R, Soares DG, et al. PARPs and the DNA damage response. Carcinogenesis 2012;33:1433-40. [PubMed]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev 2012;26:417-32. [PubMed]
- Byun TS, Pacek M, Yee MC, et al. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 2005;19:1040-52. [PubMed]
- Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J 2012;443:13-26. [PubMed]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003;300:1542-8. [PubMed]
- Liu S, Shiotani B, Lahiri M, et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell 2011;43:192-202. [PubMed]
- Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol 2002;22:555-66. [PubMed]
- Kim JE, McAvoy SA, Smith DI, et al. Human TopBP1 ensures genome integrity during normal S phase. Mol Cell Biol 2005;25:10907-15. [PubMed]
- Kumagai A, Lee J, Yoo HY, et al. TopBP1 activates the ATR-ATRIP complex. Cell 2006;124:943-55. [PubMed]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev 2002;16:198-208. [PubMed]
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 2001;276:47759-62. [PubMed]
- Wang J, Gong Z, Chen J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol 2011;193:267-73. [PubMed]
- Gong Z, Kim JE, Leung CC, et al. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell 2010;37:438-46. [PubMed]
- Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol 2009;184:793-804. [PubMed]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A 2003;100:13827-32. [PubMed]
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA. PLoS Biol 2003;1:E33 [PubMed]
- Delacroix S, Wagner JM, Kobayashi M, et al. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 2007;21:1472-7. [PubMed]
- Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell 2010;21:926-35. [PubMed]
- Mordes DA, Glick GG, Zhao R, et al. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 2008;22:1478-89. [PubMed]
- Brush GS, Morrow DM, Hieter P, et al. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci U S A 1996;93:15075-80. [PubMed]
- Liu S, Bekker-Jensen S, Mailand N, et al. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 2006;26:6056-64. [PubMed]
- Block WD, Yu Y, Lees-Miller SP. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res 2004;32:997-1005. [PubMed]
- Guo Z, Kumagai A, Wang SX, et al. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev 2000;14:2745-56. [PubMed]
- Liu Q, Guntuku S, Cui XS, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev 2000;14:1448-59. [PubMed]
- Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 2006;40:209-35. [PubMed]
- Gotter AL, Suppa C, Emanuel BS. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol 2007;366:36-52. [PubMed]
- Nedelcheva MN, Roguev A, Dolapchiev LB, et al. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J Mol Biol 2005;347:509-21. [PubMed]
- Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci U S A 2006;103:18143-7. [PubMed]
- Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem 2007;282:2729-40. [PubMed]
- Unsal-Kaçmaz K, Chastain PD, Qu PP, et al. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol 2007;27:3131-42. [PubMed]
- Smith KD, Fu MA, Brown EJ. Tim-Tipin dysfunction creates an indispensible reliance on the ATR-Chk1 pathway for continued DNA synthesis. J Cell Biol 2009;187:15-23. [PubMed]
- Errico A, Costanzo V, Hunt T. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc Natl Acad Sci U S A 2007;104:14929-34. [PubMed]
- Kondratov RV, Antoch MP. Circadian proteins in the regulation of cell cycle and genotoxic stress responses. Trends Cell Biol 2007;17:311-7. [PubMed]
- Kemp MG, Akan Z, Yilmaz S, et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem 2010;285:16562-71. [PubMed]
- Chini CC, Chen J. Human claspin is required for replication checkpoint control. J Biol Chem 2003;278:30057-62. [PubMed]
- Lee J, Kumagai A, Dunphy WG. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol Cell 2003;11:329-40. [PubMed]
- Jeong SY, Kumagai A, Lee J, et al. Phosphorylated claspin interacts with a phosphate-binding site in the kinase domain of Chk1 during ATR-mediated activation. J Biol Chem 2003;278:46782-8. [PubMed]
- Walker M, Black EJ, Oehler V, et al. Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene 2009;28:2314-23. [PubMed]
- Niida H, Katsuno Y, Banerjee B, et al. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol Cell Biol 2007;27:2572-81. [PubMed]
- Smits VA, Reaper PM, Jackson SP. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol 2006;16:150-9. [PubMed]
- Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol 2001;13:738-47. [PubMed]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009;9:153-66. [PubMed]
- Sørensen CS, Syljuasen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res 2012;40:477-86. [PubMed]
- Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 2003;3:421-9. [PubMed]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004;432:316-23. [PubMed]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 2000;14:397-402. [PubMed]
- Takai H, Tominaga K, Motoyama N, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev 2000;14:1439-47. [PubMed]
- Xu Y, Ashley T, Brainerd EE, et al. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev 1996;10:2411-22. [PubMed]
- Jallepalli PV, Lengauer C, Vogelstein B, et al. The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J Biol Chem 2003;278:20475-9. [PubMed]
- Zachos G, Rainey MD, Gillespie DA. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J 2003;22:713-23. [PubMed]
- Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev 2009;23:2394-9. [PubMed]
- Bansbach CE, Betous R, Lovejoy CA, et al. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 2009;23:2405-14. [PubMed]
- Yusufzai T, Kong X, Yokomori K, et al. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 2009;23:2400-4. [PubMed]
- Ciccia A, Bredemeyer AL, Sowa ME, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 2009;23:2415-25. [PubMed]
- Postow L, Woo EM, Chait BT, et al. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 2009;284:35951-61. [PubMed]
- Ghosal G, Yuan J, Chen J. The HARP domain dictates the annealing helicase activity of HARP/SMARCAL1. EMBO Rep 2011;12:574-80. [PubMed]
- Yuan J, Ghosal G, Chen J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol Cell 2012;47:410-21. [PubMed]
- Weston R, Peeters H, Ahel D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev 2012;26:1558-72. [PubMed]
- Ciccia A, Nimonkar AV, Hu Y, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell 2012;47:396-409. [PubMed]
- Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 2008;117:219-33. [PubMed]
- Luke-Glaser S, Luke B, Grossi S, et al. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J 2010;29:795-805. [PubMed]
- Unk I, Hajdu I, Blastyak A, et al. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9:257-67. [PubMed]
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res 2009;37:3475-92. [PubMed]
- Blastyák A, Pinter L, Unk I, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 2007;28:167-75. [PubMed]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 2010;11:208-19. [PubMed]
- Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells 2008;26:5-11. [PubMed]
- Klarer AC, McGregor W. Replication of damaged genomes. Crit Rev Eukaryot Gene Expr 2011;21:323-36. [PubMed]
- Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer 2011;11:96-110. [PubMed]
- Makridakis NM, Reichardt JK. Translesion DNA polymerases and cancer. Front Genet 2012;3:174. [PubMed]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science 1999;283:1001-4. [PubMed]
- Ogi T, Shinkai Y, Tanaka K, et al. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci U S A 2002;99:15548-53. [PubMed]
- Johnson RE, Washington MT, Haracska L, et al. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature 2000;406:1015-9. [PubMed]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 2005;74:317-53. [PubMed]
- Waters LS, Minesinger BK, Wiltrout ME, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 2009;73:134-54. [PubMed]
- Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8:461-9. [PubMed]
- Hoege C, Pfander B, Moldovan GL, et al. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002;419:135-41. [PubMed]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 2003;425:188-91. [PubMed]
- Bienko M, Green CM, Crosetto N, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 2005;310:1821-4. [PubMed]
- Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle 2004;3:1011-3. [PubMed]
- Kannouche P, Fernandez de Henestrosa AR, Coull B, et al. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J 2002;21:6246-56. [PubMed]
- Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 2010;465:951-5. [PubMed]
- Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 2010;141:255-67. [PubMed]
- Broomfield S, Hryciw T, Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res 2001;486:167-84. [PubMed]
- Watanabe K, Tateishi S, Kawasuji M, et al. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 2004;23:3886-96. [PubMed]
- Motegi A, Liaw HJ, Lee KY, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci U S A 2008;105:12411-6. [PubMed]
- Masuda Y, Suzuki M, Kawai H, et al. En bloc transfer of polyubiquitin chains to PCNA in vitro is mediated by two different human E2-E3 pairs. Nucleic Acids Res 2012;40:10394-407. [PubMed]
- Davies AA, Huttner D, Daigaku Y, et al. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell 2008;29:625-36. [PubMed]
- Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem 2006;281:32081-8. [PubMed]
- Yang XH, Shiotani B, Classon M, et al. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev 2008;22:1147-52. [PubMed]
- Niimi A, Brown S, Sabbioneda S, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci U S A 2008;105:16125-30. [PubMed]
- Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res 2008;18:162-73. [PubMed]
- Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J 2011;436:527-36. [PubMed]
- Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev 2007;21:879-85. [PubMed]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell 2007;129:665-79. [PubMed]
- Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol 2006;16:586-90. [PubMed]
- Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev 2003;17:64-76. [PubMed]
- Guo C, Sonoda E, Tang TS, et al. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell 2006;23:265-71. [PubMed]
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 2004;14:491-500. [PubMed]
- Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev 2004;14:113-9. [PubMed]
- Guo C, Tang TS, Bienko M, et al. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol 2006;26:8892-900. [PubMed]
- Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell 2005;18:499-505. [PubMed]
- Fujii S, Fuchs RP. Interplay among replicative and specialized DNA polymerases determines failure or success of translesion synthesis pathways. J Mol Biol 2007;372:883-93. [PubMed]
- Centore RC, Yazinski SA, Tse A, et al. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol Cell 2012;46:625-35. [PubMed]
- Machida Y, Kim MS, Machida YJ. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle 2012;11:3395-402. [PubMed]
- Ghosal G, Leung JW, Nair BC, et al. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J Biol Chem 2012;287:34225-33. [PubMed]
- Juhasz S, Balogh D, Hajdu I, et al. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res 2012;40:10795-808. [PubMed]
- Mosbech A, Gibbs-Seymour I, Kagias K, et al. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat Struct Mol Biol 2012;19:1084-92. [PubMed]
- Davis EJ, Lachaud C, Appleton P, et al. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol 2012;19:1093-100. [PubMed]
- Kim MS, Machida Y, Vashisht AA, et al. Regulation of error-prone translesion synthesis by Spartan/C1orf124. Nucleic Acids Res 2013;41:1661-8. [PubMed]
- Povlsen LK, Beli P, Wagner SA, et al. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat Cell Biol 2012;14:1089-98. [PubMed]
- Friedberg EC. Reversible monoubiquitination of PCNA: A novel slant on regulating translesion DNA synthesis. Mol Cell 2006;22:150-2. [PubMed]
- Van Sloun PP, Varlet I, Sonneveld E, et al. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol Cell Biol 2002;22:2159-69. [PubMed]
- Hicks JK, Chute CL, Paulsen MT, et al. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol 2010;30:1217-30. [PubMed]
- Sharma S, Hicks JK, Chute CL, et al. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic Acids Res 2012;40:682-91. [PubMed]
- Okada T, Sonoda E, Yoshimura M, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol 2005;25:6103-11. [PubMed]
- Wu F, Lin X, Okuda T, et al. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res 2004;64:8029-35. [PubMed]
- Roos WP, Tsaalbi-Shtylik A, Tsaryk R, et al. The translesion polymerase Rev3L in the tolerance of alkylating anticancer drugs. Mol Pharmacol 2009;76:927-34. [PubMed]
- Sharma S, Shah NA, Joiner AM, et al. DNA polymerase zeta is a major determinant of resistance to platinum-based chemotherapeutic agents. Mol Pharmacol 2012;81:778-87. [PubMed]
- Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 2012;26:1393-408. [PubMed]
- Meetei AR, de Winter JP, Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet 2003;35:165-70. [PubMed]
- MacKay C, Declais AC, Lundin C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 2010;142:65-76. [PubMed]
- Kratz K, Schopf B, Kaden S, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 2010;142:77-88. [PubMed]
- Liu T, Ghosal G, Yuan J, et al. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 2010;329:693-6. [PubMed]
- Smogorzewska A, Desetty R, Saito TT, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 2010;39:36-47. [PubMed]
- de Laat WL, Appeldoorn E, Jaspers NG, et al. DNA structural elements required for ERCC1-XPF endonuclease activity. J Biol Chem 1998;273:7835-42. [PubMed]
- Enzlin JH, Scharer OD. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J 2002;21:2045-53. [PubMed]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem 2008;77:259-87. [PubMed]
- Chen XB, Melchionna R, Denis CM, et al. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell 2001;8:1117-27. [PubMed]
- Wang AT, Sengerová B, Cattell E, et al. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev 2011;25:1859-70. [PubMed]
- Fekairi S, Scaglione S, Chahwan C, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 2009;138:78-89. [PubMed]
- Svendsen JM, Smogorzewska A, Sowa ME, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 2009;138:63-77. [PubMed]
- Muñoz IM, Hain K, Declais AC, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 2009;35:116-27. [PubMed]
- Andersen SL, Bergstralh DT, Kohl KP, et al. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell 2009;35:128-35. [PubMed]
- Wang AT, Sengerova B, Cattell E, et al. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev 2011;25:1859-70. [PubMed]
- Knipscheer P, Raschle M, Smogorzewska A, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 2009;326:1698-701. [PubMed]
- Räschle M, Knipscheer P, Enoiu M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 2008;134:969-80. [PubMed]
- Papadopoulo D, Guillouf C, Mohrenweiser H, et al. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc Natl Acad Sci U S A 1990;87:8383-7. [PubMed]
- Niedzwiedz W, Mosedale G, Johnson M, et al. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell 2004;15:607-20. [PubMed]
- Mirchandani KD, McCaffrey RM, D’Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7:902-11. [PubMed]
- Kim H, Yang K, Dejsuphong D, et al. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol 2012;19:164-70. [PubMed]
- Ali AM, Pradhan A, Singh TR, et al. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood 2012;119:3285-94. [PubMed]
- Moldovan GL, Madhavan MV, Mirchandani KD, et al. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol 2010;30:1088-96. [PubMed]
- O’Driscoll M. Diseases associated with defective responses to DNA damage. Cold Spring Harb Perspect Biol 2012;4:a012773 [PubMed]
- Colnaghi R, Carpenter G, Volker M, et al. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin Cell Dev Biol 2011;22:875-85. [PubMed]
- Biton S, Barzilai A, Shiloh Y. The neurological phenotype of ataxia-telangiectasia: solving a persistent puzzle. DNA Repair (Amst) 2008;7:1028-38. [PubMed]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071-8. [PubMed]
- Jeppesen DK, Bohr VA, Stevnsner T. DNA repair deficiency in neurodegeneration. Prog Neurobiol 2011;94:166-200. [PubMed]
- DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol 2012;132:785-96. [PubMed]
- Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet 2008;9:321-45. [PubMed]
- Byrnes GB, Southey MC, Hopper JL. Are the so-called low penetrance breast cancer genes, ATM, BRIP1, PALB2 and CHEK2, high risk for women with strong family histories? Breast Cancer Res 2008;10:208. [PubMed]
- Spry M, Scott T, Pierce H, et al. DNA repair pathways and hereditary cancer susceptibility syndromes. Front Biosci 2007;12:4191-207. [PubMed]
- Colas C, Coulet F, Svrcek M, et al. Lynch or not Lynch? Is that always a question? Adv Cancer Res 2012;113:121-66. [PubMed]
- Hall MJ, Reid JE, Burbidge LA, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer 2009;115:2222-33. [PubMed]
- Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer 2007;7:937-48. [PubMed]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 2010;11:196-207. [PubMed]
- Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010;42:410-4. [PubMed]
- Gage M, Wattendorf D, Henry LR. Translational advances regarding hereditary breast cancer syndromes. J Surg Oncol 2012;105:444-51. [PubMed]
- Masutani C, Kusumoto R, Yamada A, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 1999;399:700-4. [PubMed]
- Kunkel TA, Pavlov YI, Bebenek K. Functions of human DNA polymerases eta, kappa and iota suggested by their properties, including fidelity with undamaged DNA templates. DNA Repair (Amst) 2003;2:135-49. [PubMed]
- Stary A, Kannouche P, Lehmann AR, et al. Role of DNA polymerase eta in the UV mutation spectrum in human cells. J Biol Chem 2003;278:18767-75. [PubMed]
- Pan Q, Fang Y, Xu Y, et al. Down-regulation of DNA polymerases kappa, eta, iota, and zeta in human lung, stomach, and colorectal cancers. Cancer Lett 2005;217:139-47. [PubMed]
- Ceppi P, Novello S, Cambieri A, et al. Polymerase eta mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res 2009;15:1039-45. [PubMed]
- Albertella MR, Lau A, O’Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst) 2005;4:583-93. [PubMed]
- Tan XH, Zhao M, Pan KF, et al. Frequent mutation related with overexpression of DNA polymerase beta in primary tumors and precancerous lesions of human stomach. Cancer Lett 2005;220:101-14. [PubMed]
- Yoshizawa K, Jelezcova E, Brown AR, et al. Gastrointestinal hyperplasia with altered expression of DNA polymerase beta. PLoS One 2009;4:e6493 [PubMed]
- Lemée F, Bergoglio V, Fernandez-Vidal A, et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A 2010;107:13390-5. [PubMed]
- Palles C, Cazier JB, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 2013;45:136-44. [PubMed]
- Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer 2012;12:587-98. [PubMed]
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer 2012;12:801-17. [PubMed]
- Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004;303:844-8. [PubMed]
- Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med 2004;10:1321-8. [PubMed]
- Cheok CF, Verma CS, Baselga J, et al. Translating p53 into the clinic. Nat Rev Clin Oncol 2011;8:25-37. [PubMed]
- Bykov VJ, Issaeva N, Shilov A, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med 2002;8:282-8. [PubMed]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [PubMed]
- Panda H, Jaiswal AS, Corsino PE, et al. Amino acid Asp181 of 5'-flap endonuclease 1 is a useful target for chemotherapeutic development. Biochemistry 2009;48:9952-8. [PubMed]
- Gao Z, Maloney DJ, Dedkova LM, et al. Inhibitors of DNA polymerase beta: activity and mechanism. Bioorg Med Chem 2008;16:4331-40. [PubMed]
- Chen X, Zhong S, Zhu X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res 2008;68:3169-77. [PubMed]
- Samol J, Ranson M, Scott E, et al. Safety and tolerability of the poly(ADP-ribose) polymerase (PARP) inhibitor, olaparib (AZD2281) in combination with topotecan for the treatment of patients with advanced solid tumors: a phase I study. Invest New Drugs 2012;30:1493-500. [PubMed]
- Madhusudan S, Smart F, Shrimpton P, et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res 2005;33:4711-24. [PubMed]
- Mohammed MZ, Vyjayanti VN, Laughton CA, et al. Development and evaluation of human AP endonuclease inhibitors in melanoma and glioma cell lines. Br J Cancer 2011;104:653-63. [PubMed]
- Bhaskara S, Chyla BJ, Amann JM, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 2008;30:61-72. [PubMed]
- Bhaskara S, Knutson SK, Jiang G, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010;18:436-47. [PubMed]
- Evers B, Drost R, Schut E, et al. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin Cancer Res 2008;14:3916-25. [PubMed]
- Rottenberg S, Jaspers JE, Kersbergen A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A 2008;105:17079-84. [PubMed]
- Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 2010;28:2512-9. [PubMed]
- Gelmon KA, Tischkowitz M, Mackay H, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 2011;12:852-61. [PubMed]
- McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 2006;66:8109-15. [PubMed]
- Sultana R, McNeill DR, Abbotts R, et al. Synthetic lethal targeting of DNA double-strand break repair deficient cells by human apurinic/apyrimidinic endonuclease inhibitors. Int J Cancer 2012;131:2433-44. [PubMed]
- Weston VJ, Oldreive CE, Skowronska A, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood 2010;116:4578-87. [PubMed]
- Williamson CT, Muzik H, Turhan AG, et al. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther 2010;9:347-57. [PubMed]
- Vilar E, Bartnik CM, Stenzel SL, et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res 2011;71:2632-42. [PubMed]
- Kennedy RD, Chen CC, Stuckert P, et al. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest 2007;117:1440-9. [PubMed]
- Chen CC, Kennedy RD, Sidi S, et al. CHK1 inhibition as a strategy for targeting Fanconi Anemia (FA) DNA repair pathway deficient tumors. Mol Cancer 2009;8:24. [PubMed]
- Kuriyama I, Mizuno T, Fukudome K, et al. Effect of dehydroaltenusin-C12 derivative, a selective DNA polymerase alpha inhibitor, on DNA replication in cultured cells. Molecules 2008;13:2948-61. [PubMed]
- Maeda N, Kokai Y, Ohtani S, et al. Anti-tumor effects of dehydroaltenusin, a specific inhibitor of mammalian DNA polymerase alpha. Biochem Biophys Res Commun 2007;352:390-6. [PubMed]
- Kumamoto-Yonezawa Y, Sasaki R, Suzuki Y, et al. Enhancement of human cancer cell radiosensitivity by conjugated eicosapentaenoic acid - a mammalian DNA polymerase inhibitor. Int J Oncol 2010;36:577-84. [PubMed]
- Mizushina Y, Manita D, Takeuchi T, et al. The inhibitory action of kohamaic acid A derivatives on mammalian DNA polymerase beta. Molecules 2009;14:102-21. [PubMed]
- Stachelek GC, Dalal S, Donigan KA, et al. Potentiation of temozolomide cytotoxicity by inhibition of DNA polymerase beta is accentuated by BRCA2 mutation. Cancer Res 2010;70:409-17. [PubMed]
- Yang J, Parsons J, Nicolay NH, et al. Cells deficient in the base excision repair protein, DNA polymerase beta, are hypersensitive to oxaliplatin chemotherapy. Oncogene 2010;29:463-8. [PubMed]
- Mizushina Y, Motoshima H, Yamaguchi Y, et al. 3-O-methylfunicone, a selective inhibitor of mammalian Y-family DNA polymerases from an Australian sea salt fungal strain. Mar Drugs 2009;7:624-39. [PubMed]
- Xie K, Doles J, Hemann MT, et al. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci U S A 2010;107:20792-7. [PubMed]
- Doles J, Oliver TG, Cameron ER, et al. Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A 2010;107:20786-91. [PubMed]


