
Is liquid biopsy ready for the litmus test and what has been achieved so far to deal with pre-analytical issues?
Introduction
Will there ever be a world where people are not afflicted with cancer? Not very likely, as biological processes like growth, differentiation and aging are part of life, and these processes are inextricably prone to error. Even the effective repair systems in our bodies are unable to cope with and eradicate all the genetic and epigenetic changes due to metabolism or the damage caused by external factors. Therefore, the only way to reduce the cancer burden in the long term is by detecting tumors as early as possible, preferably at a stage where the cancer is still localized and can be treated effectively and curatively. The requirements for early cancer detection (not to mention screening) are very high in terms of sensitivity and specificity. Over the last few years, we have seen an impressive progress in the development of methods aimed at the detection and quantification of tumor-associated alterations. They include, but are not limited to, real-time polymerase chain reaction (PCR), digital PCR, beam, emulsion, amplification and magnetics (BEAMing), and next generation sequencing.
Until only a few years ago, physicians relied solely on pathological analysis of tumor tissue obtained by way of a biopsy. Yet recent, findings have shown that the examination of extracellular nucleic acids—released from tumor cells into the blood stream or other body fluids—can be a useful tool in the care of cancer patients (1). This approach, named “liquid biopsy”, comprises not only the analysis of extracellular/cell-free nucleic acids but of circulating tumor cells as well. In contrast to performing a “real” biopsy, the approach of a liquid biopsy or better liquid profiling is minimally invasive—a simple blood draw only—and allows more frequent analyses of cell-free nucleic acids. Additionally, the latter is not only less expensive, but also less stressful for the patient. Thus, the analysis of alterations in cell-free DNA is possible in almost real-time. This approach will not only be useful for the care of patients already diagnosed with cancer, but can also be used in diagnostic settings or even as a screening tool in a high-risk population.
The concept of liquid biopsy has a long history and goes back several decades to when Mandel et al. for the first time described the presence of extracellular nucleic acids in humans (2). About 20 years later, the group of Stroun et al. took up on these results and were finally able to demonstrate the release of genetic material from tumor cells into the cellular environment (3). The fact that tumor-associated genetic and epigenetic alterations are present not only in plasma and serum but all body fluids, was the starting point for a development that culminated last year in the Food and Drug Administration (FDA) approval of a test able to detect mutations in the epidermal growth factor receptor (EGFR) gene in DNA not isolated from tissue but from plasma (which had been the standard procedure so far). Thanks to impressive technological developments during the last 10 years, it is now possible to monitor patients in real-time for their response to antitumor therapy. However, as impressive as these results are, the biological basis for this newly developed technology is far from rock-solid. Researchers’ understanding of the release mechanisms of nucleic acids into the environment is incomplete. Apart from necrosis and apoptosis, in vitro experiments demonstrated that DNA is released by active mechanisms not associated with cell death (4-6). Also, we still do not know which intrinsic and extrinsic factors influence the release mechanisms. This lack of knowledge makes it difficult to compare the results obtained in different laboratories. Notwithstanding the fact that several research groups all over the world have shown that the analysis of cell-free nucleic acids can be a useful method for diagnosis, therapy monitoring, early detection of remission, and the detection of therapy failure in cancer patients, a standardized approach is needed. This applies above all to the pre-analytic handling of blood samples used in liquid biopsies. For many of the factors which possibly have an influence on the quantity and quality of cell-free DNA, the available data are scarce. These factors include, but are not limited to: time of the day for blood draw, patient conditions [fasting vs. eating, liquid intake vs. not, sitting vs. laying, sex, age, body mass index (BMI)], drug use, nutritional supplements, exercise, smoking, tourniquet use for blood draw, blood collection set, blood tube, fill level of blood tube (below vs. nominal fill) and others. In this short review, we will summarize the published data on pre-analytic considerations, blood draw, influence of delayed processing of blood samples, and storage of plasma/serum. We will focus on DNA only and will not mention results obtained with RNA.
Influence of physical exercise on cell-free DNA
An increase in the amount of cell-free DNA induced by exercise has been described especially in patients whose physical fitness is below-average (7). This holds true for a single bout of high-intensive strength training like weight lifting (8) and endurance exercises like running a marathon (9). So far, it is not clear whether and to what extent this issue is relevant for the care of cancer patients.
Tourniquet use and time of blood draw
The effect of tourniquet use on cell-free DNA was analyzed in one paper and no difference was found when the amount of nucleosomes containing 5-methylcytosine or the histone modification H3K9Me3 (10). In the same paper it was also demonstrated that the time of blood draw had no influence on the number of nucleosomes containing 5-methylcytosine or the histone modification H3K9Me3.
Choice of tubes for blood draw
The majority of data on pre-analytical considerations published so far deal with choosing the optimal tubes for blood draw and the time of plasma/serum preparation (Table 1).
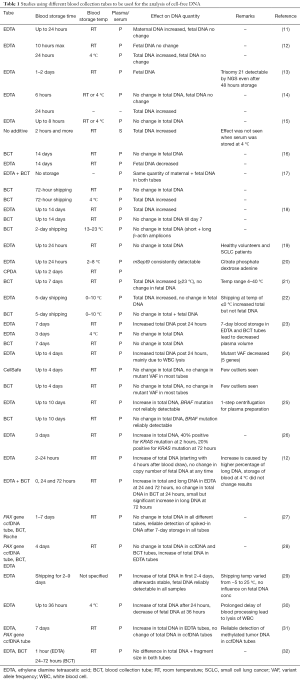
Full table
When ethylene diamine tetraacetic acid (EDTA) blood was stored for an extended period of time, it became obvious that the concentration of cell-free DNA was diluted by an increased amount of genomic DNA. This was shown to be caused by dying white blood cells (WBCs) which release their DNA into the environment. In order to prolong the time between blood draw and plasma preparation, several groups tried to stabilize WBCs and to avoid cell lysis. In the first paper by Dhallan et al., formaldehyde was added to EDTA blood and the percentage of cell-free fetal DNA (cffDNA) was compared to untreated EDTA blood (33). The researchers found an increase in the percentage of cffDNA in 7/10 blood samples (collected at one site) when formaldehyde was added (it is not clear from the paper how much time passed between blood draw and plasma preparation). In a second phase of the study, treated and untreated EDTA blood samples were collected at multiple sites and within 24 hours shipped to a central lab for processing. In all 69 formaldehyde-treated blood samples, an increased percentage of cffDNA could be observed (mean 25%). In contrast, when formaldehyde was added to EDTA blood and processed within 1 hour after blood draw, no change in the total amount of cell-free DNA or cffDNA was observed (34). A similar observation was made when formaldehyde-stabilized blood samples were processed not later than 24 hours after blood draw (35). In another report, the addition of neutral-buffered formaldehyde to EDTA blood for up to 36 hours and storage at 4 °C before plasma processing lead to a stabilization of blood cells and no “contamination” of cffDNA with genomic DNA was observed. When samples were processed within 6 hours after blood draw, the addition of formaldehyde had no effect (30). Additionally, formaldehyde inhibited a nuclease-mediated DNA degradation. Taken together, it is fair to assume that the addition of formaldehyde prevents the lysis of WBCs and therefore protects cell-free DNA, especially when there is an extended time period between blood draw and plasma preparation. These observations lead to the development of blood collection tubes (BCTs; Streck, NE, USA) specifically designed to preserve the original proportion and integrity of cffDNA in maternal plasma, and allowed the collection of blood at multiple sites and shipping at ambient temperature to a laboratory for analysis (16). Since it is known that the treatment of DNA with formaldehyde induces different alterations, Das et al. treated DNA with the reagent included in the Streck BCT tubes for up to 14 days and analyzed its effects in comparison to formaldehyde and glutaraldehyde. They demonstrated that the BCT reagent had no effect on DNA amplification by PCR while the treatment with the other two agents lead to a significant decrease in DNA amplification (36). When the quantity and fragment size distribution (as a quality marker) of cfDNA obtained in EDTA, BCT and CellSafe tubes and processed within 1 hour was compared no differences were observed (24). This holds true when blood was stored for up to 96 hours in BCT or CellSafe tubes.
In addition to Streck, several other companies (e.g., Qiagen, Roche, Biomatrica) also developed BCTs. Some of the tubes mentioned above have been tested for the detection of tumor-associated alterations in cell-free DNA from cancer patients, while for others tests are still pending (Table 1).
In a recently published paper, a strong correlation between different BCTs and the concentration of cffDNA was found (37). Additionally, it was found that the concentration of cffDNA was significantly reduced in serum compared to plasma. This is probably caused by a genomic DNA release by WBCs during clotting.
Plasma vs. serum
The question whether plasma or serum is a better substrate for the analysis of cell-free DNA cannot yet be answered unequivocally. While the DNA concentration is higher in serum than in plasma, this is probably caused by dying WBC during clot formation (38,39). Vallée et al. compared plasma and serum for the detection of EGFR mutations and found a higher detection rate in plasma (40). A similar observation was made for the detection of BRAFV600E mutation in melanoma patients (41). When plasma and serum were used for the detection of KRAS mutation by digital droplet PCR in pancreatic cancer patients, both body liquids worked equally well (42). Additionally, the use of serum for sensitive detection of mutations in KRAS, TP53 or SMAD4 with digital PCR has been described (43). The detection of BRAF mutations in melanoma patients was also possible in both plasma and serum, but the percentage of tumor-derived mutant DNA was approximately twice as high in plasma (39,44). The detection of cell-free nucleosomes modified by methylation was also equally possible in plasma and serum (10). In a meta-analysis, the detection frequencies of EGFR mutations in non-small cell lung cancer patients were compared and it was concluded that plasma or serum can be used as a surrogate for tissue (45). When the endogenous nuclease activity was measured, a 14.9-fold higher activity was found in serum compared to EDTA plasma. While a DNAse I treatment did not alter the cffDNA yields in EDTA-plasma (very likely due to the inhibiting effect of EDTA in the plasma), a complete degradation was seen in serum. The addition of increasing doses of EDTA to non-anticoagulated plasma and serum resulted in a stepwise inhibition of their nuclease activity (46). The observation that the inhibition constant of EDTA was the same in plasma and serum led us to conclude that the inhibitory effect followed the same principle in both media and that the packaging structures preventing the DNA from being degraded are similar. When Wistar rats grafted with the human colon cancer cell line SW480 were treated with a mix of DNAse I and proteases a decrease in DNA and proteins and an antitumor effect was seen (47). These observations point to an association between proteins and extracellular DNA but whether these structures are different in plasma and serum is unclear so far.
Influence of transit temperature
Transporting blood drawn into BCT tubes at 4 °C lead to an increase in the total amount of cell-free DNA but did not affect the quantity of cffDNA. This effect was not observed when the tubes were shipped at room temperature (17). Similar results were described by Wang and coworkers who shipped EDTA blood at temperatures below 0 °C for 2 days. This resulted in an increase in total DNA but not in cffDNA. When BCT tubes were used, only a moderate increase in total cell-free DNA was seen while the cffDNA concentration did not change (22).
In another report it was shown that storage/transport of maternal DNA drawn into BCT tubes for up to 7 days at temperatures of 23 °C or below did not change the concentration of cffDNA, while exposure to higher temperatures (up to 40 °C) lead to an increase of the total amount of cell-free DNA but not the cffDNA (21).
Plasma preparation
One or multiple centrifugations in combination with filtration (0.2 µM filter) can be used to separate WBC from plasma (48), but these days most research groups apply a 2-step centrifugation protocol. In the first step, a low-speed centrifugation is applied. After that, the plasma supernatant is carefully transferred into a new tube, and in the second step then centrifuged at high speed. In one paper, a minimum of two centrifugation steps is recommended and if necessary a third high-speed spin before plasma storage or DNA extraction can be added (49). An interesting observation was published by Swinkels and coworkers who demonstrated that in order to make plasma cell-free, a high-speed centrifugation (16,000 ×g) can be applied either before storage or after thawing of the frozen samples (14,50). This could be important for multicenter studies in which plasma samples are collected at different sites, frozen, and then shipped to a central laboratory for analysis. According to the above findings, all tubes could thus be subjected to high-speed centrifugation after thawing and before analysis, to make sure that plasma samples are free of cells and are treated the same way.
Storage conditions
Only a few papers have been published on the influence of storage conditions on cell-free DNA, and these give conflicting results. Storage of plasma for 2 weeks at −80 °C had no effect on the total DNA concentration according to one publication (14). When plasma was stored for more than 12 months at −80 °C, a decrease of DNA levels was observed and a BRAFV600E mutation failed to be found in a repeated test. In contrast, when DNA isolated from plasma or serum was stored at −20 °C for less than 12 months results were reproducible (41). Lee et al. stored maternal serum at −20 °C for up to 40 months and observed a decay rate of −0.66 genome equivalents/mL/month of storage for cffDNA (51). This effect was not seen in amniotic fluid which was treated in the same way. Koide and coworkers used amplicons of different sizes (63 to 524 bp length) to quantify the amount of cffDNA in plasma which had been stored for 4 years at −20 °C. They found a fragmentation similar to that of unstored samples (52). In contrast to a modest decrease in cell-free DNA described above, the yearly decay rate of 30% was much higher when plasma from lung cancer patients was stored at −80 °C for a median of 23.4 months (53). A similarly strong decrease (approx. 30%) was observed when purified DNA was stored at −20 °C for a prolonged period of time. The storage of plasma at −80 °C for 5 to 21 months resulted in a decrease of total cell-free DNA by 38%. This effect was not seen when isolated DNA was stored at −80 °C (54). Holdenrieder et al. measured the amount of nucleosomes in two series of EDTA stabilized serum samples which were stored at −70 °C (55). The samples stored for 6 to 9 months had a nucleosome concentration of 354 ng/mL, whereas samples stored for 64 months after collection had a quantity of 220 ng/mL, corresponding to a median decay of 32%.
Thawing conditions
The influence of thawing plasma/serum (fast vs. slow, optimal temperature) on the amount or integrity of cell-free DNA has not been analyzed thoroughly so far and in most reports thawing temperature is not even stated. In the instructions for the commercially available Epi proColon 2.0 kit (Epigenomics, Berlin, Germany), it is recommended to thaw frozen plasma for about 30 min at 15 to 30 °C while van Dessel et al. thawed plasma samples at 4 °C (24). Several authors recommend freezing plasma/serum in small aliquots in order to minimize freeze/thawing cycles (14,56).
DNA isolation
Page et al. compared four commercial kits for the isolation of cell-free DNA from plasma and showed that the Circulating Nucleic Acids kit (Qiagen, Hilden, Germany) gave the highest quantities (57). Similar results were published elsewhere (58). When isolation of cffDNA was performed manually with the DP virus kit (Qiagen, Germany) and compared with the isolation by an automated system (COBAS AmpliPrep DNA/RNA extractor, Roche Diagnostics, Switzerland), it was found that the former method yielded a higher amount of DNA (59). Recently, the SPIDIA program (standardization and improvement of generic pre-analytic tools and procedures for in-vitro diagnostics) performed a survey on the influence of storage conditions and extraction methods on the quantity and quality of cell-free DNA (60). Based on the DNA yields, they assort the kits in different groups, classifying them as “generic” (optimized for extraction of nucleic acids from tissue), “suitable” (allowing the recovery of circulating cell-free DNA and other nucleic acids), and “dedicated” (specifically made for the isolation of circulating cell-free DNA from plasma/serum). In their final conclusions, the authors recommend the use of dedicated kits for the isolation of cell-free DNA from body liquids.
Conclusions
From the data summarized above we believe it is fair to draw the following conclusions:
- For many of the factors which potentially have an effect on the quantity and quality of cell-free DNA, there are almost no data available. This applies, most importantly, to the factors which need to be considered before the actual blood draw;
- In addition to EDTA tubes which need to be rapidly processed after blood draw, there are new tubes from several suppliers available which stabilize cell-free DNA and prevent cell lysis. This allows a prolonged storage and facilitates shipping/transport at ambient temperature between the clinic and a laboratory. During shipping, blood samples should not be exposed to extreme temperatures (i.e., below 0 °C and higher than 30 °C) but this issue needs more data;
- In most laboratories a 2-step centrifugation (low and high speed) for a complete removal of cells is performed;
- The data on the optimal storage conditions for plasma/serum are scarce and give contradictory results. For long-term storage, ultra-low temperatures (i.e., −80 °C or lower) might be better suited. So far, it is not clear whether purified cell-free DNA is more stable and better suited for long-term storage than plasma/serum;
- The use of dedicated kits (manual or automated systems) for DNA isolation is preferable, especially when downstream applications need a high amount of DNA.
During the past few years, the very first steps towards establishing standard operating procedures for the handling of liquid biopsy samples have been taken, but more data is needed in order to build a solid basis for future guidelines.
Acknowledgments
We want to thank the numerous patients who supported our studies by donating blood, the physicians and nurses at UKH Halle/Saale, and last but not least Dana Reinicke for her invaluable help with the lab work.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.12.04). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez l'homme. C R Acad Sci Paris 1948;142:241-3.
- Stroun M, Anker P, Maurice P, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989;46:318-22. [Crossref] [PubMed]
- Anker P, Stroun M, Maurice PA. Spontaneous extracellular synthesis of DNA released by human blood lymphocytes. Cancer Res 1976;36:2832-9. [PubMed]
- Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphoctyes as shown in an in vitro system. Cancer Res 1975;35:2375-82. [PubMed]
- Bronkhorst AJ, Wentzel JF, Aucamp J, et al. Characterization of the cell-free DNA released by cultured cancer cells. Biochim Biophys Acta 2016;1863:157-65. [Crossref] [PubMed]
- Breitbach S, Tug S, Simon P. Circulating cell-free DNA: an up-coming molecular marker in exercise physiology. Sports Med 2012;42:565-86. [Crossref] [PubMed]
- Atamaniuk J, Vidotto C, Kinzlbauer M, et al. Cell-free plasma DNA and purine nucleotide degradation markers following weightlifting exercise. Eur J Appl Physiol 2010;110:695-701. [Crossref] [PubMed]
- Atamaniuk J, Stuhlmeier KM, Vidotto C, et al. Effects of ultra-marathon on circulating DNA and mRNA expression of pro- and anti-apoptotic genes in mononuclear cells. Eur J Appl Physiol 2008;104:711-7. [Crossref] [PubMed]
- Rasmussen L, Herzog M, Romer E, et al. Pre-analytical variables of circulating cell-free nucleosomes containing 5-methylcytosine DNA or histone modification H3K9Me3. Scand J Clin Lab Invest 2016;76:448-53. [Crossref] [PubMed]
- Angert RM, LeShane ES, Lo YM, et al. Fetal cell-free plasma DNA concentrations in maternal blood are stable 24 hours after collection: analysis of first- and third-trimester samples. Clin Chem 2003;49:195-8. [Crossref] [PubMed]
- Barrett AN, Zimmermann BG, Wang D, et al. Implementing prenatal diagnosis based on cell-free fetal DNA: accurate identification of factors affecting fetal DNA yield. PLoS One 2011;6:e25202 [Crossref] [PubMed]
- Buysse K, Beulen L, Gomes I, et al. Reliable noninvasive prenatal testing by massively parallel sequencing of circulating cell-free DNA from maternal plasma processed up to 24h after venipuncture. Clin Biochem 2013;46:1783-6. [Crossref] [PubMed]
- Chan KC, Yeung SW, Lui WB, et al. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin Chem 2005;51:781-4. [Crossref] [PubMed]
- Jung M, Klotzek S, Lewandowski M, et al. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem 2003;49:1028-9. [Crossref] [PubMed]
- Fernando MR, Chen K, Norton S, et al. A new methodology to preserve the original proportion and integrity of cell-free fetal DNA in maternal plasma during sample processing and storage. Prenat Diagn 2010;30:418-24. [PubMed]
- Hidestrand M, Stokowski R, Song K, et al. Influence of temperature during transportation on cell-free DNA analysis. Fetal Diagn Ther 2012;31:122-8. [Crossref] [PubMed]
- Norton SE, Lechner JM, Williams T, et al. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem 2013;46:1561-5. [Crossref] [PubMed]
- Board RE, Williams VS, Knight L, et al. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci 2008;1137:98-107. [Crossref] [PubMed]
- Distler J, Tetzner R, Weiss G, et al. Evaluation of Different Blood Collection Tubes and Blood Storage Conditions for the Preservation and Stability of Cell-Free Circulating DNA for the Analysis of the Methylated mSEPT9 Colorectal Cancer Screening Marker. Adv Exp Med Biol 2016;924:175-8. [Crossref] [PubMed]
- Wong D, Moturi S, Angkachatchai V, et al. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin Biochem 2013;46:1099-104. [Crossref] [PubMed]
- Wang Q, Cai Y, Brady P, et al. Real-time PCR evaluation of cell-free DNA subjected to various storage and shipping conditions. Genet Mol Res 2015;14:12797-804. [Crossref] [PubMed]
- Parpart-Li S, Bartlett B, Popoli M, et al. The Effect of Preservative and Temperature on the Analysis of Circulating Tumor DNA. Clin Cancer Res 2017;23:2471-7. [Crossref] [PubMed]
- van Dessel LF, Beije N, Helmijr JC, et al. Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions. Mol Oncol 2017;11:295-304. [Crossref] [PubMed]
- Denis MG, Knol AC, Theoleyre S, et al. Efficient Detection of BRAF Mutation in Plasma of Patients after Long-term Storage of Blood in Cell-Free DNA Blood Collection Tubes. Clin Chem 2015;61:886-8. [Crossref] [PubMed]
- Sherwood JL, Corcoran C, Brown H, et al. Optimised Pre-Analytical Methods Improve KRAS Mutation Detection in Circulating Tumour DNA (ctDNA) from Patients with Non-Small Cell Lung Cancer (NSCLC). PLoS One 2016;11:e0150197 [Crossref] [PubMed]
- Alidousty C, Brandes D, Heydt C, et al. Comparison of Blood Collection Tubes from Three Different Manufacturers for the Collection of Cell-Free DNA for Liquid Biopsy Mutation Testing. J Mol Diagn 2017;19:801-4. [Crossref] [PubMed]
- Warton K, Yuwono NL, Cowley MJ, et al. Evaluation of Streck BCT and PAXgene Stabilised Blood Collection Tubes for Cell-Free Circulating DNA Studies in Plasma. Mol Diagn Ther 2017;21:563-70. [Crossref] [PubMed]
- Clausen FB, Jakobsen TR, Rieneck K, et al. Pre-Analytical Conditions in Non-Invasive Prenatal Testing of Cell-Free Fetal RHD. PLoS One 2013;8:e76990 [Crossref] [PubMed]
- Zhang Y, Li Q, Hui N, et al. Effect of formaldehyde treatment on the recovery of cell-free fetal DNA from maternal plasma at different processing times. Clin Chim Acta 2008;397:60-4. [Crossref] [PubMed]
- Schmidt B, Reinicke D, Reindl I, et al. Liquid biopsy - Performance of the PAXgene(R) Blood ccfDNA Tubes for the isolation and characterization of cell-free plasma DNA from tumor patients. Clin Chim Acta 2017;469:94-8. [Crossref] [PubMed]
- Markus H, Contente-Cuomo T, Liang WS, et al. Evaluation of pre-analytical factors affecting plasma DNA analysis. Available online: https://www.biorxiv.org/content/early/2017/04/18/126839
- Dhallan R, Au WC, Mattagajasingh S, et al. Methods to increase the percentage of free fetal DNA recovered from the maternal circulation. JAMA 2004;291:1114-9. [Crossref] [PubMed]
- Chinnapapagari SK, Holzgreve W, Lapaire O, et al. Treatment of maternal blood samples with formaldehyde does not alter the proportion of circulatory fetal nucleic acids (DNA and mRNA) in maternal plasma. Clin Chem 2005;51:652-5. [Crossref] [PubMed]
- Chung GT, Chiu RW, Chan KC, et al. Lack of dramatic enrichment of fetal DNA in maternal plasma by formaldehyde treatment. Clin Chem 2005;51:655-8. [Crossref] [PubMed]
- Das K, Fernando MR, Basiaga S, et al. Effects of a novel cell stabilizing reagent on DNA amplification by PCR as compared to traditional stabilizing reagents. Acta Histochem 2014;116:55-60. [Crossref] [PubMed]
- Wong D, Moturi S, Angkachatchai V, et al. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin Biochem 2013;46:1099-104. [Crossref] [PubMed]
- Lee TH, Montalvo L, Chrebtow V, et al. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001;41:276-82. [Crossref] [PubMed]
- Aung KL, Donald E, Ellison G, et al. Analytical validation of BRAF mutation testing from circulating free DNA using the amplification refractory mutation testing system. J Mol Diagn 2014;16:343-9. [Crossref] [PubMed]
- Vallée A, Marcq M, Bizieux A, et al. Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients. Lung Cancer 2013;82:373-4. [Crossref] [PubMed]
- Daniotti M, Vallacchi V, Rivoltini L, et al. Detection of mutated BRAFV600E variant in circulating DNA of stage III-IV melanoma patients. Int J Cancer 2007;120:2439-44. [Crossref] [PubMed]
- Ako S, Nouso K, Kinugasa H, et al. Utility of serum DNA as a marker for KRAS mutations in pancreatic cancer tissue. Pancreatology 2017;17:285-90. [Crossref] [PubMed]
- Jackson JB, Choi DS, Luketich JD, et al. Multiplex Preamplification of Serum DNA to Facilitate Reliable Detection of Extremely Rare Cancer Mutations in Circulating DNA by Digital PCR. J Mol Diagn 2016;18:235-43. [Crossref] [PubMed]
- Gonzalez-Cao M, Mayo-de-Las-Casas C, Molina-Vila MA, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res 2015;25:486-95. [Crossref] [PubMed]
- Wu Y, Liu H, Shi X, et al. Can EGFR mutations in plasma or serum be predictive markers of non-small-cell lung cancer? A meta-analysis. Lung Cancer 2015;88:246-53. [Crossref] [PubMed]
- Barra GB, Santa Rita TH, de Almeida Vasques J, et al. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin Biochem 2015;48:976-81. [Crossref] [PubMed]
- Trejo-Becerril C, Perez-Cardenas E, Gutierrez-Diaz B, et al. Antitumor Effects of Systemic DNAse I and Proteases in an In Vivo Model. Integr Cancer Ther 2016;15:NP35-43. [Crossref] [PubMed]
- Chiu RW, Poon LL, Lau TK, et al. Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma. Clin Chem 2001;47:1607-13. [PubMed]
- Page K, Powles T, Slade MJ, et al. The importance of careful blood processing in isolation of cell-free DNA. Ann N Y Acad Sci 2006;1075:313-7. [Crossref] [PubMed]
- Swinkels DW, Wiegerinck E, Steegers EA, et al. Effects of blood-processing protocols on cell-free DNA quantification in plasma. Clin Chem 2003;49:525-6. [Crossref] [PubMed]
- Lee T, LeShane ES, Messerlian GM, et al. Down syndrome and cell-free fetal DNA in archived maternal serum. Am J Obstet Gynecol 2002;187:1217-21. [Crossref] [PubMed]
- Koide K, Sekizawa A, Iwasaki M, et al. Fragmentation of cell-free fetal DNA in plasma and urine of pregnant women. Prenat Diagn 2005;25:604-7. [Crossref] [PubMed]
- Sozzi G, Roz L, Conte D, et al. Effects of prolonged storage of whole plasma or isolated plasma DNA on the results of circulating DNA quantification assays. J Natl Cancer Inst 2005;97:1848-50. [Crossref] [PubMed]
- Pinzani P, Salvianti F, Orlando C, et al. Circulating cell-free DNA in cancer. Methods Mol Biol 2014;1160:133-45. [Crossref] [PubMed]
- Holdenrieder S, Von Pawel J, Nagel D, et al. Long-term stability of circulating nucleosomes in serum. Anticancer Res 2010;30:1613-5. [PubMed]
- El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013;424:222-30. [Crossref] [PubMed]
- Page K, Guttery DS, Zahra N, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 2013;8:e77963 [Crossref] [PubMed]
- Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014;406:6499-512. [Crossref] [PubMed]
- Ordoñez E, Rueda L, Cañadas MP, et al. Evaluation of sample stability and automated DNA extraction for fetal sex determination using cell-free fetal DNA in maternal plasma. Biomed Res Int 2013;2013:195363 [Crossref] [PubMed]
- Malentacchi F, Pizzamiglio S, Verderio P, et al. Influence of storage conditions and extraction methods on the quantity and quality of circulating cell-free DNA (ccfDNA): the SPIDIA-DNAplas External Quality Assessment experience. Clin Chem Lab Med 2015;53:1935-42. [Crossref] [PubMed]


