
Bioinformatics-based identification of differentiated expressed microRNA in esophageal squamous cell carcinoma
Introduction
Esophageal squamous cell carcinoma (ESCC), accounts for approximately 90% of cases of esophageal cancer worldwide (1), with 5-year survival rate declining from 20% to 4% in the advanced cases (2). Such a high mortality rate is partially attributed to late diagnosis, few therapeutic options and poor response to treatment (3,4). Therefore, the identification of new biomarkers for the early detection of ESCC and development of new target-directed therapies are urgently needed.
MicroRNA (miRNAs) is a class of small, highly conserved noncoding RNAs that negatively regulate gene expression through mRNA degradation or translational inhibition via 3'-untranslated region (3'-UTR) of the modulated gene (5). A given miRNA may have one or multiple target genes and a given gene may be regulated by several miRNAs of the same or different sequence. These short nucleotides can also be over or under expressed, promoting or impeding tumor formation based on the downstream target genes of miRNAs controlled. The miRNA expression profiles of esophageal cancer are significantly different from that of adjacent normal tissue. Several studies have demonstrated that miRNA is related to both the formation and development of EC (6). Hsa-miR-21 is found to be upregulated in squamous cell carcinoma tumor tissues and play a role in abnormal proliferation (7). Hsa-miR-203 is one of the down-regulated miRNAs in esophageal cancer and mediates the post-transcriptional suppression of SOCS3 (8). Analysis of miRNAs also has strong potential for the identification of novel prognostic or predictive biomarkers. For instance, miRNA profiles could use as the prediction of resistance to neoadjuvant radio-chemotherapy in squamous cell carcinoma of the esophagus, which indicated the miRNA are involved in the therapeutic response in EC and suggest that miRNA profiles could facilitate pre-therapeutic patient selection (9).
Although exhaustive miRNA profiling was performed, only a limited number of esophageal cancer associated miRNAs were reported (10-14). All these studies were restricted by the number of clinical samples available (less or equal to 100 tumors and matched nearby normal tissue samples). Thus, to overcome this limitation and to provide a comprehensive landscape of microRNA expression in esophageal cancer, we carried out a pooled-analysis using four public available datasets (12,15-17) comprising 675 patient samples including 349 ESCC cases and 326 normal esophageal tissues (NE). Our analysis discovers 9 new miRNAs. We add the rank of two miRNA to gene prediction to find the target genes.
Methods
Database search and collection strategy
Four datasets E-GEOD-13937, E-GEOD-43732, E-GEOD-6188 and E-GEOD-55856 were downloaded from The European Bioinformatics Institute (EMBL-EBI), ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/), according to following criteria: (I) the number of patients in each study is greater than 50; (II) tumor should company with corresponding normal esophageal tissue from the same patient; (III) all raw data must be available for download and process prior to further analysis; (IV) to reduce bias, dataset generated from the same detection platform was preferred; (V) each patient sample is unique and no duplicated samples were allowed in the analysis.
Annotation
The latest version of human miRNA database (release 21) is downloaded from the mirBase website (18) (http://www.mirbase.org/). A modification was also performed to create a new annotation reference by removing the hyphens in the original miRNA name, i.e., hsa-miR-6859-5p to hsamir68595p. For each dataset, the miRNA name is extracted from the annotation file, if necessary, it will be modified: (I) p1 occurs at the end of the miRNA name, then it is substituted by p; (II) the character ‘*’ occurs at the end of the miRNA name, then it is replaced by -3p. Each modification is then modified again and match with the miRNA name referred to. If matched with more than one entry of the reference, the miRNA name is removed. The match numbers of miRNA for each dataset is summarized in Table 1.
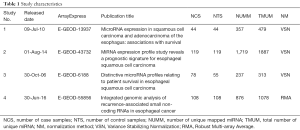
Full table
Statistical analysis
Statistical analysis is performed by R (version 3.4.3) software (19). An additional software package (ArrayExpress) was downloaded from Bioconductor (20). The within dataset normalization is carried out using Robust Multi-array Average (RMA) (21) for two-color microarray and Variance Stabilizing Normalization (VSN) (22) for single color microarray as suggested by ArrayExpress. The normalization method for each dataset is summarized in Table 1.
Each miRNA can be included in one or more studies. For each miRNA, we find out which studies have this miRNA, e.g., hsa-mir-617 and hsa-mir-1180 can be found in study 2 and 4. Then we group the miRNA according to the combination of studies, e.g., hsa-mir-617 and hsa-mir-1180 is in the same study group (2,4). If the study group has only one study, it will apply to surrogate variable analysis (SVA) by 3 surrogate variables, representing age, smoking and alcohol consumption. If the study group has more than one study, the miRNAs is normalized by Combat (23) to adjust the unknown batch effect, and then further adjusted using SVA by 3 surrogate variables. The association is calculated using one-way ANOVA, where the dependent variable is the normalized expression level of one miRNA and explanatory variables is a binary variable, where 0 is control and 1 is ESCC case. The ranked-ANOVA is also used, and the mean difference of rank between ANOVA and ranked-ANOVA is smaller than 3 among the top 50 ranked miRNAs. ANOVA has an advantage of estimating the effect size, and hence chosen in the final analysis.
Results
Study characteristics
Four miRNA microarray datasets with more than 300 patient samples were selected for this study (Table 1). All samples are matched tumor and normal. A small subset of data in both E-GEOD-13937 and E-GEOD-6188 dataset was derived from esophageal adenocarcinoma (EAC) and was excluded from this study. Only data collected from ESCC cancers and the adjacent normal tissues were subjected to further investigation. A total of 349 ESCC cases and 326 normal esophageal tissue samples (NE) were covered in this pooled analysis.
Known differentially expressed miRNAs in ESCC
Several recent lines of evidence suggested that miRNAs expression profiling could serve as both diagnostic and prognostic biomarkers in ESCC. We found 20 previously discovered miRNA in ESCC as known miRNAs. Figure 1 shows the volcano plot of the known differentially expressed miRNAs, and these miRNAs are roughly symmetrical distributed according to the positive or negative effect value. A summary table of the known miRNAs can be found in Table 2. There are 10 miRNAs with more abundant expression in adjacent normal tissues than in ESCC tissues, while there are 10 miRNAs with smaller expression in adjacent normal tissues than in ESCC tissues.
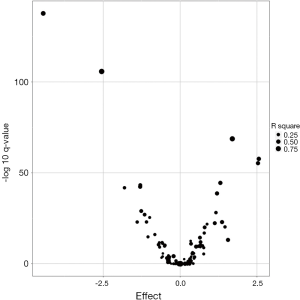
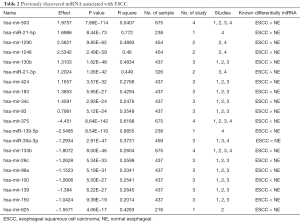
Full table
Upregulated miRNAs in ESCC
Many oncogenic miRNAs have been found to be upregulated in our study, e.g., hsa-mir-503, hsa-mir-21-5p, hsa-mir-1290, hsa-mir-1246. Among them, hsa-mir-503 is the most significant upregulated miRNA whose expression was knowingly downregulated in esophagus carcinoma cells (24). The second significantly upregulated one is hsa-mir-21-5p and the one ranking fifth is another mature body of hsa-mir-21, the hsa-mir-21-3p. Previous studies have also confirmed that hsa-mir-21 is predominantly up-regulated in ESCC (25). The third significantly upregulated miRNA is hsa-mir-1290, which has been confirmed expression upregulated in ESCC (26).
Downregulated miRNAs in ESCC
Table 2 also shows some of the miRNAs which were significantly downregulated in ESCC. Among them, hsa-mir-375 was the most significantly downregulated miRNA confirmed by previous studies (27). The second and third significantly down-regulated miRNAs are hsa-mir-139 and hsa-mir-30a respectively, and both miRNA are confirmed by previous studies in ESCC (10,28).
Previously unknown differentially expressed miRNAs in ESCC
MiRNA not mentioned as significant in published paper is considered as unknown miRNAs. It is because of these miRNAs are tested but not known as associated with ESCC. Table 3 shows the top 9 significantly unknown differentially expressed miRNAs. Only one of them was significantly down-regulated while the rest were significantly up-regulated. The first and second significantly up-regulated unknown miRNAs are hsa-mir-1180 and hsa-mir-663b respectively. The only down-regulated unknown miRNA was hsa-mir-617, which none of the studies mentioned significantly downregulated expressed in ESCC.
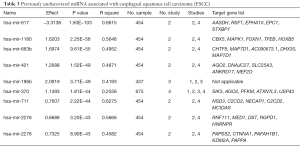
Full table
With the top 3 predicted target genes for each unknown differentially expressed miRNAs, we apply TopGo to find that GO term positive regulation of cell proliferation (GO:0008284) is the most significant enriched GO term (Table 4).
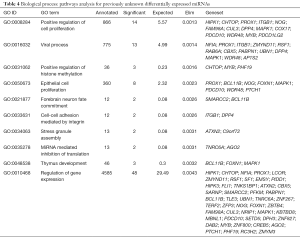
Full table
MiRNA target prediction strategy
The analytical strategy to predict the biological targets of ESCC associated miRNAs is to average the ranked output data from three popular miRNA database, TargetScan (29), Diana (30) and mirDB (31), followed by sorting them in ascending order. Top five target genes for each miRNA are listed in Table 3.
Discussion
Recently, a large number of differentially expressed miRNAs have been observed in esophageal cancer, which supported the value of miRNAs in diagnosis and prognosis. Some studies used qRT-PCR to study a limited number of miRNAs with up to 100 matched tumor and normal samples (7,11), and other experiments used microarrays and qRT-PCR (12-14). In our study, the combination of these four miRNAs microarrays included up to 675 samples [349 ESCC cases and 326 normal esophageal tissue samples (NE)], of which the sample size and number of patients both increased. To our knowledge, this is the most comprehensive meta-analysis to date. For known miRNAs, 95 out of 108 miRNAs can be found in more than one study.
The most significantly upregulated known miRNA was hsa-mir-503. Interestingly, many studies showed that it is down-regulated in many cancers compared with adjacent normal tissues, such as cervical cancer, NSCLC, osteosarcoma, EEC, cervical cancer and hepatocellular carcinoma (32-36). Another study also found the downregulation of miR-503 expression in ESCC tissue relative to adjacent normal tissues in ESCC (37). However our comprehensive pooled analysis of four microarrays indicated that hsa-mir-503 is an upregulated miRNA. Target gene prediction showed that the most promising targets are CCNE1 and FGF2. CCNE1 (also known as Cyclin E1) is a well-known gene associated with cell proliferation and FGF2 is a frequently mutated cancer gene in ESCC (38).
A recent publication shows that hsa-mir-503 downregulated HOXC13, by directly targeting its 3'-UTR, and inhibited proliferation of ESCC. Previous studies have also confirmed that hsa-mir-21, the second significant differentiated miRNA, is predominantly up-regulated in ESCC (28) and has shown it significantly increased in heavy drinking patients. The third significantly upregulated miRNA is hsa-mir-1290, which has been shown that the targeting site of hsa-mir-1290 is located in 3'-UTR of NFIX (29). Also the inverse correlation between the levels of hsa-mir-1290 and NFIX protein and miRNA in ESCC tissue samples were also confirmed.
Hsa-mir-375 was the most significantly downregulated miRNA confirmed by previous studies (30). It has been shown that SHOX2 is a direct target of hsa-mir-375, where SHOX2 expression is upregulated in ESCC. Further analysis showed that SHOX2 induced proliferation, invasion, and metastasis of ESCC. The second and third significantly down-regulated miRNAs is hsa-mir-139 and hsa-mir-30a respectively. It has been shown that the target of hsa-mir-139 is NR5A2, which induce cell cycle arrest in the G0/G1 phase and to suppress the invasive capability of esophageal carcinoma cells (10). MiRNA hsa-mir-30a is closely associated with advanced ESCC progression and poor prognosis of patients with ESCC by promoting ESCC cell proliferation (31).
At present, many studies have verified that the tumorigenic with hsa-mir-21 upregulation in ESCC is associated with target genes PDCD4, K-RAS, PTEN and ERK1/2/MAPK pathway (39-42). Meanwhile, a study suggested that hsa-mir-21 down-regulation suppresses cell growth, invasion and induced cell apoptosis by targeting FASL, TIMP3, and RECK genes in esophageal carcinoma (26). Several studies have found that hsa-mir-1290 promotes cancer progression by targeting nuclear factor I/X (NFIX) and SCAI in ESCC (43,44). Similarly, hsa-mir-130b plays a carcinogenic role by inhibiting PTEN expression in ESCC (45). In summary, these markedly upregulated miRNAs are predominantly tumorigenic, and they are likely to be prognostic biomarkers for ESCC Table 5.
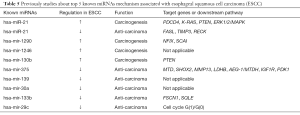
Full table
The most significantly downregulated miRNA in our study is hsa-mir-375. Accumulated evidence indicated that this microRNA rendered its strong tumor-suppressive effect through targeting at SHOX2, MMP13, LDHB, AEG-1/MTDH, IGF1R and PDK1 genes (46-50). hsa-mir-139-5p is one of miRNA previously discovered to suppress tumor growth and invasiveness in human ESCC through unknown biological targets (51) (Table 5). The target of has-mir-139a is NR5A2 and hsa-mir-30a is associated with ESCC cell proliferation.
The most significantly up-regulated miRNA is hsa-mir-1180, which may modulate the expression of predicted target gene MAPK1 and participate in a variety of cellular processes such as proliferation, differentiation, transcription regulation and development. Surprisingly, the down-regulation of this miRNA has been observed in pancreatic cancers with anti-cancer activity via regulation of the TNIP2/NF-KB signaling pathway and its downstream proteins, including MMP-2, MMP-9, Bax, Bcl-2, p21, and cyclin D1 (52).
The most significantly downregulated miRNA was hsa-mir-617, which was one of the top 9 significantly differentially expressed unknown miRNAs. To our knowledge, we are the first research group to demonstrate the reverse association of this miRNA with esophageal cancer. A negative correlation in the expression of this microRNA and tumor has also been observed in oral cavity and oral verrucous carcinoma (53). Five potential modulated genes of this microRNA are AASDH, RSF1, EPHA10, EPC1 and STXBP1. Among these targets, RSF1 was related with tumor proliferation, invasion and chemo-resistance, and EPC1 promotes cell apoptosis and DNA repair.
We have to admit limitations in this analysis. Firstly, Clinical characteristics of patients are collected from the GEO and ArrayExpress database. None of clinical characteristics of patients are present in all four studies, and hence we could not use any clinical characteristics of patients. However, we apply Surrogate Variable Analysis (SVA) to take into account most commonly known confounder variables, i.e., age, smoking and alcohol consumption. Secondly, this study focus on ESCC only. In Europe, it is preferably a sporadic disease with a stable incidence, while in some areas, like China, Iran, Pakistan, it is rather an endemic disease with a high prevalence. Thus, our analysis might not represent a genomic map of all race. Third, the “newly described” miRNA in the analysis is based on the time when that four included arrays were conducted. Some might be defined later by other studies but it still showed a “new” one here. Last, ESCC patients enrolled at different clinical centers from the United States, Canada, China and Japan, thus multi-racial study makes it impossible to reveal one racial character, but it would represent more population worldwide.
Since the first ESCC related miRNA was reported in 2006, both the samples size and number of patients in relevant studies are continually increasing. We have performed the largest meta-analysis study with individual patient data on ESCC miRNA microarrays, which provided guidance for discovering the new biomarkers of ESCC for early diagnosis and targeted therapies. The role and mechanism of these miRNAs in ESCC need to be further investigated.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (81672270); Key project of Guangzhou Science Technology and Innovation Committee (201707020042).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.10.15). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med 2014;371:2499-509. [Crossref]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref]
- Meng XR, Lu P, Mei JZ, et al. Expression analysis of miRNA and target mRNAs in esophageal cancer. Braz J Med Biol Res 2014;47:811-7. [Crossref]
- Wen SW, Zhang YF, Li Y, et al. Characterization and effects of miR-21 expression in esophageal cancer. Genet Mol Res 2015;14:8810-8. [Crossref]
- Zhang F, Yang Z, Cao M, et al. MiR-203 suppresses tumor growth and invasion and down-regulates MiR-21 expression through repressing Ran in esophageal cancer. Cancer Lett 2014;342:121-9. [Crossref]
- Gusella M, Pezzolo E, Modena Y, et al. Predictive genetic markers in neoadjuvant chemoradiotherapy for locally advanced esophageal cancer: a long way to go. Review of the literature. Pharmacogenomics J 2018;18:14-22. [Crossref]
- Kano M, Seki N, Kikkawa N, et al. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer 2010;127:2804-14. [Crossref]
- Ogawa R, Ishiguro H, Kuwabara Y, et al. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med Mol Morphol 2009;42:102-9. [Crossref]
- Guo Y, Chen Z, Zhang L, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res 2008;68:26-33. [Crossref]
- Yang H, Gu J, Wang KK, et al. MicroRNA expression signatures in Barrett's esophagus and esophageal adenocarcinoma. Clinical Cancer Research An Official Journal of the American Association for Cancer Research 2009;15:5744-52. [Crossref]
- Lee KH, Goan YG, Hsiao M, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res 2009;315:2529-38. [Crossref]
- Mathe EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res 2009;15:6192-200. [Crossref]
- Chen Z, Li J, Tian L, et al. MiRNA expression profile reveals a prognostic signature for esophageal squamous cell carcinoma. Cancer Lett 2014;350:34-42. [Crossref]
- Jang HJ, Lee HS, Burt BM, et al. Integrated genomic analysis of recurrence-associated small non-coding RNAs in oesophageal cancer. Gut 2017;66:215-25. [Crossref]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res 2004;32:D109-11. [Crossref]
- Null RDCT, Team R, Null RCT, et al. A Language and Environment for Statistical Computing. Computing 2011;1:12-21.
- Kauffmann A, Rayner TF, Parkinson H, et al. Importing ArrayExpress datasets into R/Bioconductor. Bioinformatics 2009;25:2092-4. [Crossref]
- Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref]
- Huber W, von Heydebreck A, Sultmann H, et al. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002;18:S96-104. [Crossref]
- Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882-3. [Crossref]
- Zhao K, Chen BJ, Chen ZG, et al. Effect of miR-503 Down-Regulation on Growth and Invasion of Esophagus Carcinoma and Related Immune Function. Med Sci Monit 2015;21:3564-9. [Crossref]
- Huang G. Erratum: Differential expression of miR-21 and miR-75 in esophageal carcinoma patients and its clinical implication. Am J Transl Res 2017;9:1961.
- Xie R, Wu SN, Gao CC, et al. Prognostic value of combined and individual expression of microRNA-1290 and its target gene nuclear factor I/X in human esophageal squamous cell carcinoma. Cancer Biomark 2017;20:325-31. [Crossref]
- Hu C, Lv L, Peng J, et al. MicroRNA-375 suppresses esophageal cancer cell growth and invasion by repressing metadherin expression. Oncol Lett 2017;13:4769-75. [Crossref]
- Yang M, Liu R, Sheng J, et al. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of esophageal squamous cell carcinoma. Oncol Rep 2013;29:169-76. [Crossref]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005 [Crossref]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013;41:W169-73 [Crossref]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146-52. [Crossref]
- Liu L, Qu W, Zhong Z. Down-regulation of miR-503 expression predicate advanced mythological features and poor prognosis in patients with NSCLC. International Journal of Clinical & Experimental Pathology 2015;8:5609.
- Chong Y, Zhang J, Guo X, et al. MicroRNA-503 acts as a tumor suppressor in osteosarcoma by targeting L1CAM. Plos One 2014;9:e114585 [Crossref]
- Xu YY, Wu HJ, Ma HD, et al. MicroRNA-503 suppresses proliferation and cell-cycle progression of endometrioid endometrial cancer by negatively regulating cyclin D1. FEBS J 2013;280:3768-79. [Crossref]
- Xiao F, Zhang W, Chen L, et al. MicroRNA-503 inhibits the G1/S transition by downregulating cyclin D3 and E2F3 in hepatocellular carcinoma. J Transl Med 2013;11:195. [Crossref]
- Hu N, Kadota M, Liu H, et al. Genomic Landscape of Somatic Alterations in Esophageal Squamous Cell Carcinoma and Gastric Cancer. Cancer Research 2016;76:1714. [Crossref]
- Jiang L, Zhao Z, Zheng L, et al. Downregulation of miR-503 Promotes ESCC Cell Proliferation, Migration, and Invasion by Targeting Cyclin D1. Genomics Proteomics Bioinformatics 2017;15:208-17. [Crossref]
- Gu L, Zhang J, Shi M, et al. The effects of miRNA-1180 on suppression of pancreatic cancer. Am J Transl Res 2017;9:2798-806.
- Li P, Mao WM, Zheng ZG, et al. Down-regulation of PTEN expression modulated by dysregulated miR-21 contributes to the progression of esophageal cancer. Digestive Diseases & Sciences 2013;58:3483-93. [Crossref]
- Liu T, Liu Q, Zheng S, et al. MicroRNA-21 promotes cell growth and migration by targeting programmed cell death 4 gene in Kazakh's esophageal squamous cell carcinoma. Dis Markers 2014;2014:232837 [Crossref]
- Liu F, Zheng S, Liu T, et al. MicroRNA-21 promotes the proliferation and inhibits apoptosis in Eca109 via activating ERK1/2/MAPK pathway. Mol Cell Biochem 2013;381:115-25. [Crossref]
- Wang N, Zhang CQ, He JH, et al. MiR-21 down-regulation suppresses cell growth, invasion and induces cell apoptosis by targeting FASL, TIMP3, and RECK genes in esophageal carcinoma. Dig Dis Sci 2013;58:1863-70. [Crossref]
- Mao Y, Liu J, Zhang D, et al. MiR-1290 promotes cancer progression by targeting nuclear factor I/X(NFIX) in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother 2015;76:82-93. [Crossref]
- Yu T, Cao R, Li S, et al. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer 2015;15:29. [Crossref]
- Li X, Lin R, Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig Dis Sci 2011;56:2849-56. [Crossref]
- Kong KL, Kwong DL, Chan TH, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 2012;61:33-42. [Crossref]
- Isozaki Y, Hoshino I, Nohata N, et al. Identification of novel molecular targets regulated by tumor suppressive miR-375 induced by histone acetylation in esophageal squamous cell carcinoma. Int J Oncol 2012;41:985-94. [Crossref]
- Osako Y, Seki N, Kita Y, et al. Regulation of MMP13 by antitumor microRNA-375 markedly inhibits cancer cell migration and invasion in esophageal squamous cell carcinoma. Int J Oncol 2016;49:2255-64. [Crossref]
- Jing Chen. MiR-375 suppresses invasion and metastasis by direct targeting of SHOX2 in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 2017;49:159-69.
- Liu R, Yang M, Meng Y, et al. Tumor-suppressive function of miR-139-5p in esophageal squamous cell carcinoma. PLoS One 2013;8:e77068 [Crossref]
- Yin ZL, Wang YL, Ge SF, et al. Reduced expression of miR-503 is associated with poor prognosis in cervical cancer. Eur Rev Med Pharmacol Sci 2015;19:4081-5.
- Lajer CB, Nielsen FC, Friis-Hansen L, et al. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: a prospective translational study. Br J Cancer 2011;104:830-40. [Crossref]
- Deng Z, Wang Y, Fang X, et al. Research on miRNA-195 and target gene CDK6 in oral verrucous carcinoma. Cancer Gene Ther 2017;24:282-8. [Crossref]


