
CircNT5E/miR-422a: a new circRNA-based ceRNA network in glioblastoma
The vast majority of human transcriptome is composed of non-coding RNAs (ncRNAs) that contribute to genetic and epigenetic regulation through complex interactions with proteins and nucleic acids. Circular RNAs (circRNAs) are a conserved class of ncRNAs, generated from a non-canonical back splicing process, that catalyzes a covalent bond between the 5' and 3' ends of a single-stranded RNA molecule (1). The absence of free ends makes circular RNAs extremely stable and resistant to degradation. Many circRNAs have a tissue-specific expression and are particularly abundant in mammalian brain (2). Moreover, multiple evidences show that circRNAs are critical in cancer pathogenesis exhibiting both oncogenic and tumor suppressive properties (3). However, despite their high expression level, the biological functions of circRNAs are still poorly understood. Interestingly, a critical function assigned to circRNAs is the ability to act as “competing endogenous RNAs” (ceRNAs), subtracting microRNAs from their natural mRNA targets. As a result, circRNAs regulate gene expression establishing a dynamic crosstalk with numerous transcripts (4).
Glioblastoma (GBM) is the most common and aggressive primary malignant brain tumor in adults. The standard therapeutic options include surgery, radiation and chemotherapy. Despite a better understanding of GBM pathogenesis, tumor heterogeneity, localization and invasiveness still limit the overall survival (12–15 months) (5,6). The advances in next generation sequencing (NGS) have recently revealed the dysregulation of numerous ncRNAs in GBM. Specifically, several reports have demonstrated the oncogenic and the oncosuppressive role of miRNAs in this tumor type (7). In this context, the authors have highlighted the tumor suppressive potential of miR-422a (8). In particular, the analysis of the open database miRNA map 2.0 showed miR-422a enrichment in brain tissues while, experimentally, miR-422a was found downmodulated in malignant GBM tumors. In addition, evaluation of 381 GBM samples of The Cancer Genome Atlas (TCGA) underlined a correlation between miR-422a expression and patient survival. In functional assays, overexpression of miR-422a reduced GBM cells oncogenic properties both in vitro and in vivo. Moreover, the authors demonstrated that miR-422a directly targeted PIK3CA, a key member of the PI3K/Akt signaling pathway, which is frequently dysregulated in GBM (9,10). These findings extended to GBM the oncosuppressive role of miR-422a previously observed in multiple cancer types (11-14) and unveiled the relevance of the miR-422a/PIK3CA axis in GBM pathogenesis.
Recently, Wang et al. examined the molecular mechanism responsible for miR-422a downregulation in GBM (15). To this end, they selected 3 GBM tumors in which miR-422a was expressed at low levels. Moreover, in GBM cells they verified that miR-422a downmodulation was not due to promoter methylation. Microarray profiling on these specific GBM tumors revealed hundreds of upregulated transcripts. Through an integrated bioinformatic analysis, 38 circRNAs/lncRNAs containing two or more target sites for miR-422a were selected and subjected to a biotin-miR-422a pull-down assay to test their possible interaction with the microRNA. Among the top enriched 16 RNA candidates, a circRNA arising from the 5'-nucleotidase NT5E gene (circNT5E) exhibited the highest enrichment upon pull down and was chosen for further characterization. CircNT5E was particularly stable and localized into the cytoplasm. Interestingly, RNA in situ hybridization revealed a correlation between circNT5E expression and the clinical classification of glioma samples. In addition, circNT5E overexpression was confirmed in a cohort of 18 GBM specimens, indicating a potential oncogenic function for circNT5E in this tumor type. Accordingly, circNT5E ectopic expression in GBM cells significantly promoted proliferation, migration, invasion and tumor growth in vivo. On the other hand, circNT5E knockdown caused opposite effects both in vitro and in vivo.
Consequently, Wang et al. investigated the circNT5E biogenesis and which factors influenced its overexpression in GBM. By using a CRISPR/Cas9-based strategy, the authors removed reverse complementary matches (RCMs) from circNT5E flanking introns and observed a reduction in circNT5E expression (with no effect on its linear counterpart). This proved that the presence of intronic RCMs sequences was required for circNT5E formation, in accordance with previous studies on circRNAs biogenesis (16-18). Moreover, several works reported that RNA binding proteins, such as MBL, QKI, and ADAR can modulate circRNAs expression (19). Interestingly, a member of the ADAR family, the brain-enriched RNA-editing enzyme ADARB2 was significantly upregulated in GBM tumors at protein level, while its mRNA counterpart was substantially reduced. These controversial findings suggest a complex regulatory role for ADARB2 in GBM, which merit to be further investigated. Strikingly, ADARB2 overexpression in GBM cells increased circNT5E levels and, contemporarily, decreased the linear NT5E mRNA. In addition, RNA immunoprecipitation (RIP) and RNA pull-down assays showed a reciprocal binding between ADARB2 and NT5E pre-mRNA, demonstrating the role of ADARB2 as upstream regulator of circNT5E.
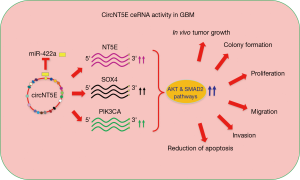
Finally, Wang et al. explored the function of circNT5E as competitive endogenous RNA. Bioinformatic analysis of circNT5E sequence revealed the presence of 20 different binding sites for miRNAs. Experimentally, miR-422a exhibited the lowest expression level upon circNT5E transfection. A luciferase reporter assay and a co-localization experiment proved the interaction between circNT5E and miR-422a. Moreover, analysis of two validated miR-422a targets (PIK3CA and NT5E) showed that circNT5E expression was sufficient to rescue their downregulation caused by miR-422a overexpression. In functional assays, miR-422a ectopic expression counteracted the oncogenic potential of circNT5E. Mechanistically, Wang et al. analyzed the circNT5E/miR-422a signaling network. NT5E, SOX4, PI3KCA, p-Akt, p-Smad2 were downregulated at protein level in miR-422a overexpressing cells or upon circNT5E knockdown. Conversely, circNT5E overexpression or miR-422a blockage resulted in the upregulation of these targets. Overall, these data indicated that circNT5E acts as a potent miR-422a sponge, playing a critical role in GBM tumorigenesis (Figure 1).
In the last few years, growing attention has been showed to the biological relevance of ncRNAs in cancer. Notably, Wang et al. detected in GBM thousands of dysregulated circRNAs/lncRNAs, whose function and potential reciprocal interaction remains practically unknown. In this context the application of an NGS approach could be extremely useful to discover additional RNA regulatory circuits. However, the discovery phase should include a larger cohort of GBM patients. Moreover, a more robust analysis should also take into consideration the genetic alterations of the analyzed tumors. This integrated approach can likely reveal unexplored and more robust targets for future translational studies. Furthermore, the implementation of more advanced bioinformatic tools can likely define novel molecular networks and identify critical targets for innovative therapies.
Interestingly, numerous transcripts were found upregulated in GBM tumors characterized by the miR-422a-downregulation. These RNAs can potentially act as additional sponges for miR-422a, thus supporting secondary ceRNA interactions. On the other hand, Wang et al. suggest that circNT5E may subtract multiple miRNAs from their targets. However, functional studies are limited to miR-422a. Generally, circRNAs with a paradigmatic sponge activity contain numerous binding sites for miRNAs (20). Since circNT5E act as an oncogene, it is highly possible that other miRNAs can be affected by its sponging activity. Thus, a more accurate analysis on its sequence and on miRNAs level in other cancer types could be useful to better define its molecular function in tumorigenesis.
Finally, the circRNAs stability and abundance in body fluids, suggest their possible use as new biomarkers in liquid biopsy, a non-invasive diagnostic technique alternative to surgical biopsies (21). In this context circNT5E, which is present at high levels in GBM tumors and correlates with clinical classification, may represent the first candidate, while the circNT5E/miR-422a network unveils novel GBM vulnerabilities.
Acknowledgments
Funding: R Taulli research is supported by
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.11.13). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205-11. [Crossref] [PubMed]
- Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018;37:555-65. [Crossref] [PubMed]
- Mitra A, Pfeifer K, Park KS. Circular RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer Res 2018;7:S624-8. [Crossref] [PubMed]
- Pearson JRD, Regad T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct Target Ther 2017;2:17040. [Crossref] [PubMed]
- Davis ME. Glioblastoma: Overview of Disease and Treatment. Clin J Oncol Nurs 2016;20:S2-8. [Crossref] [PubMed]
- Yu W, Liang S, Zhang C. Aberrant miRNAs Regulate the Biological Hallmarks of Glioblastoma. Neuromolecular Med 2018;20:452-74. [Crossref] [PubMed]
- Liang H, Wang R, Jin Y, et al. MiR-422a acts as a tumor suppressor in glioblastoma by targeting PIK3CA. Am J Cancer Res 2016;6:1695-707. [PubMed]
- Li X, Wu C, Chen N, et al. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016;7:33440-50. [PubMed]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807-12. [Crossref] [PubMed]
- Zhang J, Yang Y, Yang T, et al. Double-negative feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1 regulates hepatocellular carcinoma tumor growth and metastasis. Hepatology 2015;61:561-73. [Crossref] [PubMed]
- Faltejskova P, Svoboda M, Srutova K, et al. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med 2012;16:2655-66. [Crossref] [PubMed]
- Gougelet A, Pissaloux D, Besse A, et al. Micro-RNA profiles in osteosarcoma as a predictive tool for ifosfamide response. Int J Cancer 2011;129:680-90. [Crossref] [PubMed]
- Molina-Pinelo S, Gutiérrez G, Pastor MD, et al. MicroRNA-dependent regulation of transcription in non-small cell lung cancer. PLoS One 2014;9:e90524. [Crossref] [PubMed]
- Wang R, Zhang S, Chen X, et al. CircNT5E Acts as a Sponge of miR-422a to Promote Glioblastoma Tumorigenesis. Cancer Res 2018;78:4812-25. [Crossref] [PubMed]
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015;10:170-7. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838-47. [Crossref] [PubMed]
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272-83. [Crossref] [PubMed]
- Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res 2017;36:152. [Crossref] [PubMed]


