Expression of centrosomal protein 55 in glioma tissue and the influence on glioma cell functions
Introduction
Glioma originates from glial cells of the human nervous system, as a primary brain tumor with the highest incidence rate in humans to date (1). In China, the morbidity rate of glioma is about 10–20/100,000, and its incidence rate accounts for about 1–3% of those of systemic malignant tumors and approximately 46% of those of intracranial tumors (2). According to epidemiological studies, the median survival time of low-grade (WHO grade II) gliomas is 6–8 years, and those of high-grade (WHO grades III and IV) gliomas are only 3 years and 1 year respectively (3). At present, glioma is mainly treated through surgical resection plus postoperative radiotherapy or chemotherapy. Many new therapies for glioma have been developed, including postoperative combination chemotherapy with bevacizumab and lomustine (4), as well as postoperative stereotaxic radiological technique and high-dose radiotherapy for elderly patients with glioblastoma (5). However, these therapies can only prolong the survival of patients instead of targeting gliomas. Therefore, basic research on glioma is of great significance.
The centrosome is a non-membranous organelle as well as a crucial microtubule-organizing center in cells. It is located in the periphery of the nucleus, with a diameter of about 1–2 µm, playing pivotal roles in cell cycle and mobility. The formation of centrosomal microtubule nucleus and the anchoring action of microtubules regulate their number, stability, polarity and spatial arrangement in cells, and also dominantly maintain cell morphology and material transport. Centrosomal abnormalities may lead to dysfunction of the spindle apparatus during division and appearance of genomes with aneuploidy. These aneuploid cells and unstable genomes are closely related to the onset of malignant tumors. Meanwhile, various regulatory proteins associated with centrosomes also accumulate therein along with their maturation. Their abnormal expressions cause abnormal replication of the centrosome, directly affect microtubule tissue, induce spindle apparatus disorders and mediate the progression of cell cycle (6). Centrosomal protein 55 (CEP55) is a centrosome-related protein. Fabbro et al. (7) found that CEP55 coupled to centrosomes and intermediates, and regulated the cell cycle after being phosphorylated by ERK2 and CDK1 simultaneously. The mechanism of action was proposed by Doxsey (8). CEP55 is first phosphorylated by ERK2 and CDK1 on scaffold proteins Pent B and CG-Nap, then released into the cytoplasm and rephosphorylated from PLK1 to be fully activated, finally regulating the cell cycle. Besides, Fabbro et al. (7) reported that CEP55 gene was silenced by siRNA, using HeLa cells treated with GFP-siRNA as negative control. With elapsed time, the expression level of CEP55 was lower than that of control cells, accompanied by abnormalities of multinuclear cells and appearance of metaphase spindles (more than 2). Many cells were arrested in the intermediate phase and cytokinesis failed. Collectively, CEP55 plays key regulatory roles in spindle apparatus assembly, polarization, separation and cytokinesis. In the last decade, the roles of CEP55 in breast cancer, stomach cancer, bladder cancer, etc. have been extensively studied (9-11). Nevertheless, the influence of CEP55 on glioma still lacks systematic research. Thereby motivated, we herein detected the expressions of CEP55 in glioma tissues, and assessed the effects on glioma cell functions and apoptosis.
Methods
Glioma tissues and cells
Fifty fresh astrocytoma tissue samples resected in our hospital from May 2016 to May 2017 were collected. There were 32 males and 18 females aged 38–64 years old, with a median age of (53.5±7.6). They were diagnosed according to the WHO 2016 Classification of Gliomas. There were 26 cases of WHO grade II, 16 cases of WHO grade III and 8 cases of WHO grade IV. All patients were not treated with radiotherapy or chemotherapy before surgery, and all samples collected from surgeries were pathologically diagnosed to have astrocytomas. Meanwhile, twenty normal brain tissue samples, which were resected from patients with severe brain traumas receiving emergency decompression surgeries, were collected as a control group. This study has been approved by the ethics committee of our hospital (approval number: NJ20161209YS).
Glioma cell lines LN229, T98G, U87, U251 and HEB were purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.
Immunohistochemical assay
Fresh glioma tissue and normal control tissue samples were immediately fixed in 4% paraformaldehyde solution for 15–20 min, sectioned, embedded with paraffin, bake-dried in a 60 °C incubator for 1 h to be stuck onto a glass slide, soaked in xylene solution for 10 min twice, and hydrated with gradient concentrations of ethanol solutions (100% for 5 min, 95% for 5 min, 90% for 5 min, 85% for 5 min and 75% for 5 min). Afterwards, the sections were soaked in tap water for 3 min and then in PBS for 5 min, heated in 0.01 M boiling citrate solution by microwave for 5 min, cooled for 5 min and heated again for 5 min. After being cooled down to room temperature, the sections were rinsed with double-distilled water twice, incubated with PBS in a 37 °C humidified box for 10 min, rinsed three times with PBS (5 min each time), blocked with goat serum at 37 °C for 10 min, incubated with diluted primary antibody (1:500) at 4 °C overnight, washed three times with PBS (5 min each time), incubated with diluted biotinylated secondary antibody at 37 °C for 20 min, washed three times with PBS (5 min each time), incubated with diluted horseradish peroxidase-labeled streptavidin tertiary antibody at 37 °C for 20 min, and washed three times with PBS (5 min each time). Then freshly prepared 50 µL of DAB color development solution was dropped onto the sections for 3 min of reaction. The sections were thereafter washed with tap water for 4 min, counterstained with hematoxylin for 2 min, dehydrated with gradient concentrations of ethanol solutions, transparentized with xylene, mounted with neutral resin and observed under a light microscope. The cells with clear brownish yellow particles in the cytoplasm or nucleus were determined as positive. Ten high-power visual fields were randomly selected for each section. Percentage of positive cells to total ones: <10%, negative tissue expression; ≥10%, positive tissue expression.
Cell culture
All cells were cultured in a 37 °C incubator with 5% CO2. LN229, U87 and HEB cells were cultured in DMEM containing 10% fetal bovine serum (FBS), streptomycin (100 µg/mL) and penicillin (100 U/mL). T98G cells were cultured in MEM containing 10% FBS, streptomycin (100 µg/mL), penicillin (100 U/mL), 1% non-essential amino acids and 1 mM sodium pyruvate.
Cell transfection
Cells in the logarithmic growth phase were seeded into six-well plates at the density of 5×105 per well, and 2 mL of pre-warmed complete medium was added into each well. After 24 h of inoculation, the confluence reached about 50–60%. Then 5 µL of Lipofectamine 2000 was added into 200 µL of serum-free DMEM or MEM, gently mixed, and left still for 5 min at room temperature. Subsequently, 100 pmol siRNA was added into 200 µL of serum-free DMEM or MEM, and gently mixed. The diluted siRNA and Lipofectamine 2000 were gently mixed, and placed at room temperature for 20 min to form a siRNA/Lipofectamine 2000 complex. Then 400 µL of solution containing the siRNA/Lipofectamine 2000 complex was added into the wells containing cells and 1,600 µL of complete medium, gently shaken and mixed. After the cells were incubated for 24–48 h in a CO2 incubator at 37 °C, other experimental procedures after transfection were continued. siRNA CEP55: F: 3'-TTUUUAGAAACCAUGAAGACC-5'; R: 3'-TTGGUCUUCAUGGUUUCUAAA-5'. siRNA NC: F: 3'-TTUGCACUGUGCAAGCCUCUU-5'; R: 3'-TTAAGAGGCUUGCACAGUGCA-5'. They were synthesized by Shanghai GenePharma Co., Ltd. (China).
Western blot
Cells were collected through digestion and centrifugation, resuspended by using RIPA (50 mM pH 7.5 Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), ultrasonicated, and centrifuged at 12,000 rpm and 4 °C for 10 min. Then 50 µg of each sample was subjected to SDS-PAGE, and the products were electronically transferred to a nitrocellulose membrane. Afterwards, the membrane was blocked with 5% skimmed milk at room temperature for 1 h, incubated with primary antibody at 4 °C overnight, rinsed, and incubated with secondary antibody at 37 °C for 1 h. Scanning was conducted after ECL development. The relative expression of protein was corrected by that of internal reference and analyzed by Quantity One software.
Real-time PCR
Cells were collected through digestion and centrifugation, from which mRNA was extracted according to the instructions of mRNA extraction kit. The expression of CEP55 mRNA was detected by TaqMan mRNA detection kit, and real-time PCR data were analyzed using SYBR green II fluorescent staining and IQ5TM real-time PCR detection system (Bio-Rad, USA). The results were corrected by U6 RNA internal reference. The relative expression of CEP55 mRNA was expressed by 2−∆∆Ct, and three independent experiments were performed.
MTT assay
Cells were digested and resuspended into 5×104/mL using corresponding culture medium. The cell suspension was seeded into 96-well plates at 100 µL/well, placed in a 37 °C incubator with 5% CO2, and added MTT (5 mg/mL, 20 µL/well) at 0, 12, 24, 48, 72 h respectively for 4 h of incubation. Then culture medium in the well was removed, and the cells were added 150 µL/well of DMSO and fully mixed to detect the absorbance at 570 nm by a microplate reader.
Survival rate (%) = A570experimental group/A570control group × 100%.
Scratch assay
Five equidistant lines were drawn first on the back of each well of six-well culture plate, with each line running through the entire well. Each well was inoculated with 5×105 cells. After 24 h of culture, the confluence reached 70%. Then 2 mL of fresh complete medium was added into each well, and the cells were cultured in a 37 °C incubator for 24 h until the confluence reached 90%. After the original complete medium was discarded, a line perpendicular to the back of the well was drawn using a 10 µL tip. The cells were rinsed twice with PBS, 3 mL each time. Then 2 mL of serum-free complete medium was added into each well. The culture plate was placed in a 37 °C incubator, taken out at 0 and 24 h respectively, and placed under a microscope to observe cell migration. The changes of migration areas at the same time were calculated by ImageJ software.
Transwell assay
BD Matrigel and serum-free DMEM were diluted in a ratio of 5:1 and gently mixed. Subsequently, 100 µL of the mixture was added into the upper layer of each Transwell chamber and incubated for 4–6 h in a 37 °C incubator until Matrigel solidified. After digestion, the cells were centrifuged, resuspended in pre-warmed serum-free medium and counted. The Transwell chamber was washed three times with PBS and added the resuspended cell suspension at 100 µL per well. Complete medium containing 20% FBS was added into wells of the lower layer of culture plate. The cells were thereafter cultured in a 37 °C incubator for 12–24 h. The Transwell chamber was taken out, with the outer layer washed three times by PBS. Afterwards, the chamber was fixed in 95% ethanol solution for 10 min, and then washed by PBS. Then 0.05% crystal violet was added into each well for 10 min of staining. Subsequently, the cells were washed three times with PBS. Matrigel and the cells in the upper layer of Transwell chamber were thereafter gently wiped off with a cotton swab. The chamber was observed under an inverted microscope to count the number of cells penetrating the membrane.
Flow cytometry
Cells were digested, counted and then inoculated into a six-well plate. After 48 h of culture, the medium was discarded, and the cells were washed with PBS, digested with trypsin and resuspended with PBS. According to the instructions of annexin V kit, the cells were suspended with 500 µL of binding buffer, and then 5 µL of annexin V-FITC and 5 µL of propidium iodide were mixed for 15 min of reaction in dark at room temperature. The apoptosis was detected by flow cytometry.
Statistical analysis
All data were analyzed by SPSS19.0 software and expressed as mean ± standard deviation. Inter-group comparisons were performed by the t-test. The categorical data were subjected to the χ2 test. All images were analyzed by ImageJ software. P<0.05 was considered statistically significant.
Results
High expression of CEP55 in glioma tissue
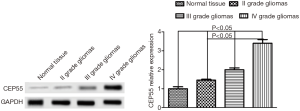
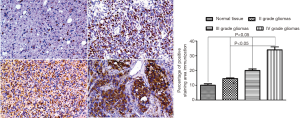
We first used Western blot to detect the expression of CEP55 protein in different grades of glioma tissues and normal brain tissues, and observed the expression by immunohistochemical staining. The expression of CEP55 protein in glioma tissues was significantly higher than that in the normal control group (P<0.05). Meanwhile, CEP55 protein expressions in different grades of gliomas also differed. The difference was not statistically significant between grade II and III gliomas, but statistically significant between grade II and IV gliomas and between grade III and grade IV gliomas (P<0.05) (Figure 1). The same results were obtained by immunohistochemical assay. In the normal control group, CEP55 protein was weakly positively stained, whereas in glioma tissues, CEP55 protein staining was strongly positive, mainly around the nucleus and cytoplasm (Figure 2).
CEP55 expressions in different cell lines
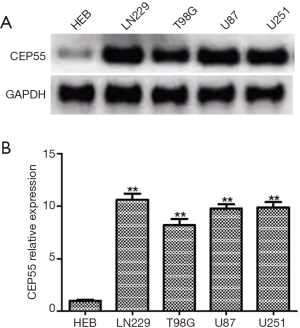
We detected the expressions of CEP55 protein in human glial cell line HEB and glioma cell lines LN229, T98G, U87 and U251 by Western blot. The expression level of CEP55 protein in human glial cells was low, but those in glioma cell lines increased to various degrees (Figure 3). Moreover, the expression level of CEP55 protein in HEB cells was significantly different from those of glioma cells.
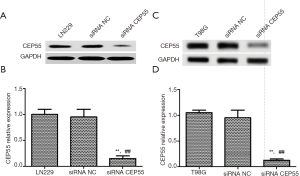
Cell transfection with siRNA for CEP55 gene expression interference
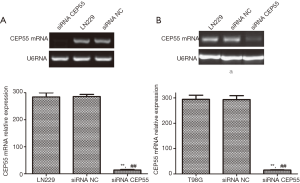
LN229 and T98G cell lines with high CEP55 protein expressions were transfected with siRNA CEP55 and siRNA NC interference fragments. The interference of CEP55 protein expression was detected by Western blot. The expression of CEP55 protein significantly decreased in the siRNA CEP55 group, which was significantly different from those of the negative control group and blank group (P<0.05), suggesting that cell lines stably silencing CEP55 had been successfully constructed (Figure 4). Consistently, RT-PCR showed that CEP55 mRNA significantly decreased in the siRNA CEP55 group, with significant difference from those of the negative control group and blank group (P<0.001) (Figure 5).
Effects of CEP55 on cell proliferation
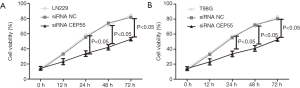
Normal LN229/T98G cells, siRNA CEP55 LN229/T98G cells and siRNA NC LN229/T98G cells were seeded at the same density in six-well culture plates. The proliferation ability was detected by MTT assay at 0, 12, 24, 48 and 72 h after inoculation. The proliferation curve of normal LN229/T98G cells was basically the same as that of siRNA NC LN229/T98G cells, whereas siRNA CEP55 LN229/T98G cells grew significantly slower than normal LN229/T98G cells and siRNA NC LN229/T98G cells did at 24, 48 and 72 h (P<0.05) (Figure 6).
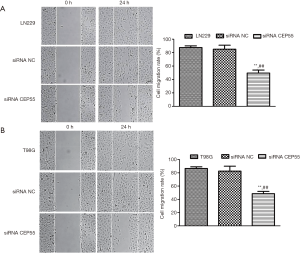
Effects of CEP55 on cell migration
Normal LN229/T98G cells, siRNA CEP55 LN229/T98G cells, and siRNA NC LN229/T98G cells were seeded at the same density in six-well culture plates. A cell-free zone was drawn using a 10 µL pipette tip. The cells that were exfoliated after scratching were taken with PBS and photographed. After 24 h of culture, photographs were taken to observe the migration ability. After normal LN229/T98G cells and siRNA NC LN229/T98G cells were cultured for 24 h, the scratch area changed greatly, and the migration was fast. However, the migration area of siRNA CEP55 LN229/T98G cells did not change much, and the migration was significantly slower than those of normal LN229/T98G cells and siRNA NC LN229/T98G cells (P<0.001) (Figure 7).
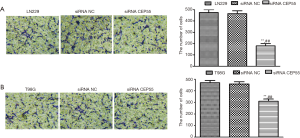
Effects of CEP55 on cell invasion
Normal LN229/T98G cells, siRNA CEP55 LN229/T98G cells and siRNA NC LN229/T98G cells at the same number were seeded in Matrigel-containing Transwell chambers, cultured under the same conditions for about 18 h, and then stained with crystal violet. The cells penetrating the chamber membrane were counted in the visual field at the same magnification. The invasive abilities of normal LN229/T98G cells and siRNA NC LN229/T98G cells were basically the same, but that of siRNA CEP55 LN229/T98G cells significantly reduced (P<0.001) (Figure 8).
Effects of CEP55 on cell apoptosis
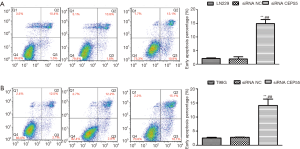
Flow cytometry exhibited that the early apoptosis rate of siRNA CEP55 LN229/T98G cells significantly exceeded those of normal LN229/T98G and siRNA NC LN229/T98G cells (P<0.001) (Figure 9).
Discussion
Glioma remains a world-class problem that plagues neurosurgery. Its onset and progression are caused by many factors, and the underlying mechanism is still unclear (12). In recent years, with the development of molecular biology, epigenetics, immunology and detection methods, more coding and non-coding genes have been closely related to gliomas, paving the way for early screening of glioma, biological immunotherapy and prognosis evaluation. Although a large number of glioma suppressor genes have been discovered so far, including coding genes p15, p21, p53, Rb, PTEM (13) and non-coding genes H19, CRNDE, miRNA-16 (14), it is still mainly treated by surgical resection and postoperative radiotherapy and chemotherapy, also with unsatisfactory prognosis (15).
As an important regulatory protein in the pericentriolar material, CEP55 dominantly participates in the assembly of cell spindles and the cleavage function of cytoplasm. CEP55 gene, composed of 464 amino acids, is located on a gene sequence of human chromosome 10q23.33 containing 9 exons (7). CEP55 can couple its C-terminus with microtubule-aggregation-related protein CG-Nap and Pent B, and regulate γ-tubulin by multiple phosphorylation sites. Thus, although CEP55 is not a protein essential for microtubule aggregation or spindle assembly, it can be mediated through a variety of proteins (16). When CEP55 protein is abnormally expressed, it can cause central structural and functional abnormalities that are common in tumor cells, indicating that CEP55 may be related with tumor occurrence (17). When the expression of CCEP55 is knocked down, the probability of cytoplasmic division failure is increased, rendering most cells unable to undergo cytokinesis, which may be one of the causes for apoptosis (18).
The expression of CEP55 is significantly higher in tissues of bladder transitional cell carcinoma than in benign prostatic hyperplasia tissues, being associated with various clinical and pathological characteristics (11). Tao et al. reported that CEP55 was highly expressed in human gastric cancer tissues and cells, also affecting cell proliferation (9). The same phenomenon has been observed in breast cancer (10). Moreover, the high expression of CEP55 has been closely related to the invasiveness and prognosis of ovarian epithelial cancer (19). In this study, Western blotting and immunohistochemical staining verified that the expression of CEP55 in glioma tissues was significantly higher than that of normal brain tissues, which was also raised with increasing WHO grade. Likewise, immunohistochemical staining suggested that the expression level of CEP55 protein may be associated with the grade of glioma. CEP55 protein also had different expression levels in different cell lines. Normal glial cells had significantly lower expression levels of CEP55 protein than those of glioma cells. Furthermore, the protein expression differences between different glioma cell lines may be ascribed to cell source and degree of malignancy. After CEP55 gene was knocked down, the proliferative, migration and invasion abilities of glioma cells were significantly lower than those of normal cells, accompanied by significantly facilitated apoptosis. Therefore, decreased expression of CEP55 can significantly inhibit the function of glioma cells and increases their apoptosis.
Conclusions
In summary, the expression of CEP55 increased in glioma tissue, and interfering with its expression significantly weakened cell functions and promoted apoptosis. Hence, CEP55 is a potential molecular marker for diagnosing gliomas or a new target for molecular therapy.
Acknowledgments
Funding: This study was financially supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.33). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all patients. The study was approved by the ethics committee of our hospital (No. NJ20161209YS).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Malzkorn B, Reifenberger G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr Opin Oncol 2016;28:494-501. [Crossref] [PubMed]
- Wang HF, Fu SL, Song JD, et al. Recent advance in marker gene of gliomas. Chin J Neuromed 2014;13:429-32.
- Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro Oncol 2013;15:ii1-56. [Crossref] [PubMed]
- Seystahl K, Wick W, Weller M. Therapeutic options in recurrent glioblastoma—an update. Crit Rev Oncol Hematol 2016;99:389-408. [Crossref] [PubMed]
- Redmond KJ, Mehta M. Stereotactic radiosurgery for glioblastoma. Cureus 2015;7:e413. [PubMed]
- Srsen V, Merdes A. The centrosome and cell proliferation. Cell Div 2006;1:26. [Crossref] [PubMed]
- Fabbro M, Zhou BB, Takahashi M, et al. Cdk1/Erk2-and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell 2005;9:477-88. [Crossref] [PubMed]
- Doxsey SJ. Molecular links between centrosome and midbody. Mol Cell 2005;20:170-2. [Crossref] [PubMed]
- Tao J, Zhi X, Tian Y, et al. CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumour Biol 2014;35:4389-99. [Crossref] [PubMed]
- Wang Y, Jin T, Dai X, et al. Lentivirus-mediated knockdown of CEP55 suppresses cell proliferation of breast cancer cells. Biosci Trends 2016;10:67-73. [Crossref] [PubMed]
- Singh PK, Srivastava AK, Rath SK, et al. Expression and clinical significance of Centrosomal protein 55 (CEP55) in human urinary bladder transitional cell carcinoma. Immunobiology 2015;220:103-8. [Crossref] [PubMed]
- Nagane M. Genetic alterations and biomarkers for glioma. Brain Nerve 2012;64:537-48. [PubMed]
- Thomas L, Di Stefano AL, Ducray F. Predictive biomarkers in adult gliomas: the present and the future. Curr Opin Oncol 2013;25:689-94. [Crossref] [PubMed]
- Yan Y, Xu Z, Li Z, et al. An insight into the increasing role of LncRNAs in the pathogenesis of gliomas. Front Mol Neurosci 2017;10:53. [Crossref] [PubMed]
- Kumthekar PU, Macrie BD, Singh SK, et al. A review of management strategies of malignant gliomas in the elderly population. Am J Cancer Res 2014;4:436-44. [PubMed]
- van der Horst A, Simmons J, Khanna KK. Cep55 stabilization is required for normal execution of cytokinesis. Cell Cycle 2009;8:3742-9. [Crossref] [PubMed]
- Kumar A, Rajendran V, Sethumadhavan R, et al. CEP proteins: the knights of centrosome dynasty. Protoplasma 2013;250:965-83. [Crossref] [PubMed]
- Martinez-Garay I, Rustom A, Gerdes HH, et al. The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics 2006;87:243-53. [Crossref] [PubMed]
- Zhang W, Niu C, He W, et al. Upregulation of centrosomal protein 55 is associated with unfavorable prognosis and tumor invasion in epithelial ovarian carcinoma. Tumour Biol 2016;37:6239-54. [Crossref] [PubMed]