
Multi-omics analysis reveals the BRCA1 mutation and mismatch repair gene signatures associated with survival, protein expression, and copy number alterations in prostate cancer
Introduction
Prostate cancer demonstrated a higher degree of heritability than other common cancers such as ovarian and breast cancer (1). Molecular profiles are increasingly being untilized for subtyping cancers and guiding treatment selection. Several studies have demonstrated the possibility in diversifying prostate cancer. One of the recent recognition is the germline and somatic mutations. Deleterious mutation in BRCA1 and BRCA2 which are associated with breast, ovarian and pancreatic cancer, have also shown to increase the risk of prostate cancer and some are even associated with aggressive form of the disease (2-4). In addition, early study from The Cancer Genome Atlas (TCGA) found that other genes, FANCD2, CKD12, and ATM classified under DNA repaired genes were also shown a higher prevalence in primary prostat cancer. It is estimated that 7–12% of men with metastatic prostate cancer involved inherited germline mutation in DNA repair genes (5).
The prevalence of somatic and germline mutations in DNA repair genes is becoming better understood. However, disease prognosis and treatment response associated with these mutations are not yet elucidated. Therapeutic management of metastatic castration-resistant prostate cancer (mCRPC) is currently based on new hormonal therapies (abiraterone, enzalutamide) and taxane-type chemotherapy (docetaxel or cabazitaxel). Early reports shown that patients carried genes associated with DNA-repair demonstrated a good response to poly-ADP ribose polymerase (PARP) inhibitors and platinum-based chemotherapy (6,7). Although results are still emerging and need validation, current evidence has suggested a potential subgroup associated with DNA repair genes mutation in prostate cancer may benefit from other treatment strategies.
Normal intracellular metabolism and environmental exposure constantly induce DNA reactivation and damage of the genome of cells in the human body. Various DNA repair mechanisms have been identified which help the cells repairing the many accidental lesions that occur continually in DNA and to protect genome integrity. Inherited and acquired deficiencies in these DNA repair mechanisms can modify cancer susceptibility as well as therapy response. The carcinogenesis of prostate cancer is suggested an accumulation of molecular changes of AR transcription activity, error prone DNA repair, oncogenic replication and changes in chromatin architecture. To further understand the role of DNA repair genes associated with other genomic alteration and signaling pathway, we applied an integrative analysis of multi-omics to TCGA prostate cancer dataset which contains 498 patients. Furthermore, we reveal the signature of “chromosome condensation”, “BRCA1”, and “mismatch repair” were associated with disease-free survival. Through the concurrent analysis of gene expression profiles, reverse phase protein lysate microarray (RPPA) data, and copy number alterations, we found the three signatures are associated with cell cycle and DNA repair pathway and also lots events of copy number alterations. This finding will support future understanding of how DNA repair processes, and DNA double-strand break repair in particular, are regulated during the cell cycle associated with the disease progression of prostate cancer.
Methods
Signature score of a gene set
To evaluate the activities of the gene sets examined in the study, the scoring method of Tian et al. (8) was used. Suppose there are N genes in a given gene set. Let xl = {x1,l, …, xN,l}, where xj,l is the log2-transformed expression level of gene j in the expression profile of sample l. For a given gene set, the signature score of a sample is defined as
where zj,l=(xj,l-μj)/σj, and μj and σj are the mean and standard deviation of the expression level of gene j in all expression profiles.
Gene sets
Total 10,236 gene sets were collected from 4 sources: Gene Ontology (GO), chemical and genetic perturbations (CGP), hallmark gene sets, and motif gene sets were used. The gene sets were downloaded from Molecular Signature database (MSigDB v3.1, http://www.broadinstitute.org/gsea/msigdb/index.jsp).
TCGA and The Cancer Protein Atlas (TCPA) data sets
Multiple genomics data of 498 prostate tumors were downloaded from the The Cancer Genome Altas (TCGA) website (http://cancergenome.nih.gov/) and also cBioPortal for Cancer Genomics (http://www.cbioportal.org/). Data includes gene expression profiles, the status of DNA copy number alternation and reverse phase protein array (RPPA) data were analyzed. The Reverse phase protein array (RPPA) data of lung adenocarcinoma datasets were downloaded from The Cancer Protein Atlas (TCPA) website (9). Data used in this study are publicly available which do not constitute research involving human subjects.
Results
Gene expression signatures of DNA mismatch repair, BRCA1 mutations, and mitosis are intercorrelated in prostate cancer
We studied the prognostic roles of biological functions (represented by gene signatures) in prostate cancer. We re-analyzed expression profiles of 498 prostate tumors of TCGA and calculated per-sample gene signature scores of curated gene sets. Signature scores of the collected 10,236 gene sets were analyzed by a Cox hazard regression. A total of 285 gene sets were significantly associated with relapse-free survival (RFS). Heatmap of signature scores of the 285 gene sets is shown in Figure 1A. Among these gene sets, 6 gene sets were associated with mitosis and 24 were associated with cell cycle. “GO CHROMOSOME CONDENSATION” was significantly associated with RFS (P=1.5×10−11). Chromosome condensation is an essential process during mitosis phase of cell cycle and thus may mark tumor progression and aggressiveness. A sub gene set “GO MITOTIC CHROMOSOME CONDENSATION”, which contains 14 out of its 31 member genes, was ranked as the 6th significant gene set (P=2.0×10−6). The results indicated that cell cycle is highly associated with cancer progression in prostate cancer. Chromosome condensation in the mitotic phase is the most prognostic gene set.
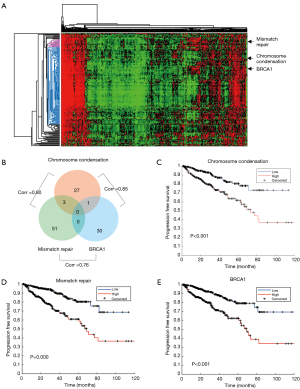
We further investigated the role of the DNA repair pathway in cancer progression. Six prognostic gene sets were correlated with DNA repair (“MATZUK MEIOTIC AND DNA REPAIR”, “GO DNA REPAIR”, “KAUFFMANN DNA REPAIR GENES”, “GO REGULATION OF DOUBLE STRAND BREAK REPAIR VIA HOMOLOGOUS RECOMBINATION”, “GO DNA SYNTHESIS INVOLVED IN DNA REPAIR”, and “GO MISMATCH REPAIR”). We also observed a BRCA1 mutation associated gene set “PUJANA BREAST CANCER WITH BRCA1 MUTATED UP” which was originally derived from differential expressed genes associated with BRCA1 mutation in breast cancer. We selected the three gene sets, “GO CHROMOSOME CONDENSATION”, “GO MISMATCH REPAIR”, and “PUJANA BREAST CANCER WITH BRCA1 MUTATED UP”, in representation of the signatures of chromosome condensation (and mitosis), mismatch repair, and BRCA1 mutation pathway for further investigation. Although member genes were rarely overlapped among the three gene sets (Figure 1B), their signature scores were highly correlated (Figure 1B). The results indicate the functional cooperation and interplay among these functions. Kaplan–Meier plots showed that patients with higher signature score of the gene set had worse prognosis (Figure 1C,D,E).
Investigations of member genes of the three prognostic gene signatures of mitosis and DNA repair
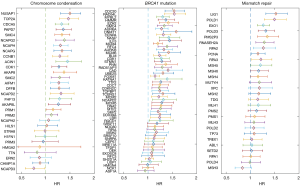
There were 30, 55, and 28 member genes of the “chromosome condensation”, “mismatch repair” and “BRCA1 mutation” gene sets, respectively. Cox regression showed that 61% (19/31), 69% (38/55), and 32% (10/31) of them had significant adverse effects in RFS [hazard ratio (HR) >0 and P<0.05; Figure 2]. Of note, NCAPD3 of the “chromosome condensation” signature was the only with significant protective hazard ratio (HR=0.76; Figure 2). In the “chromosome condensation” signature, NUSAP1, a coding gene of a nucleolar-spindle-associated protein, had the highest HR (HR=1.50) and TOP2A, a well-studied oncogene that encodes a DNA topoisomerase, was the second (HR=1.47). The two genes have been reported with roles in chromosome condensation and mitosis (10,11). The top 2 genes of the “BRCA1 mutation” signature were CDC20 and RAD54L. CDC20 is a protein of the anaphase-promoting complex which could inhibit BRCA1 facilitated homologous recombination (12). RAD54L is involved in the BRCA1 mediated homologous recombination repair (13). In the “mismatch repair” gene set, LIG1 (1st ranked) is DNA Ligase 1 that functions in DNA replication and recombination. POLD1 (DNA polymerase delta 1) also plays a critical role in DNA replication and repair (14). Overall, analysis of member genes brought biologically meaningful results that supported our findings at the gene signature level.
Proteomics validation of the activities of mitotic and DNA repair pathways
At both the gene and gene set levels, our data suggested that DNA repair and the BRCA1 pathway are prognostic and correlated with mitosis. We used the RPPA data of TCPA (9) to confirm such findings in the protein level. We calculated correlation coefficients between a signature score and proteomic activities of its members. Proteins with high correlation coefficients are showed in Table 1. The correlation coefficients of Cyclin B1 are 0.675 (ranked 1st), 0.651 (ranked 2nd), 0.495 (ranked 1st) to the three signatures, respectively. Cyclin B1 has been reported as a regulatory protein involved in mitosis. Chek2, a serine-threonine kinase involved in DNA repair, was correlated with both “chromosome condensation” and “BRCA1 mutation” signatures. Our data also showed Brca2 was negatively correlated to “mismatch repair” signature (coefficient =−0.317). The results again confirmed the association of BCRA1 and DNA repair pathways with mitosis.
Table 1
| Gene set | Protein | Corr | Gene set | Protein | Corr | Gene set | Protein | Corr |
|---|---|---|---|---|---|---|---|---|
| Chromosome condensation | Cyclin B1 | 0.675 | BRCA1 mutation | Cyclin B1 | 0.651 | Mismatch repair | Cyclin B1 | 0.497 |
| TFRC | 0.454 | TFRC | 0.468 | Chk2 | 0.370 | |||
| Caspase-7cleavedD198 | 0.385 | RBM15 | 0.423 | ACC_pS79 | 0.326 | |||
| PCNA | 0.375 | Syk | 0.419 | Bim | 0.325 | |||
| Chk2 | 0.359 | Bim | 0.404 | XRCC1 | 0.320 | |||
| Rictor | −0.330 | DJ-1 | −0.392 | PKC-alpha_pS657 | −0.301 | |||
| ACVRL1 | −0.349 | ACVRL1 | −0.426 | PKC-alpha | −0.315 | |||
| PKC-alpha_pS657 | −0.355 | PKC-alpha_pS657 | −0.428 | BRCA2 | −0.317 | |||
| Caveolin-1 | −0.356 | PKC-alpha | −0.432 | ACVRL1 | −0.353 | |||
| PKC-alpha | −0.358 | Caveolin-1 | −0.445 | Caveolin-1 | −0.353 |
TCGA, The Cancer Genome Atlas; TCPA, The Cancer Protein Atlas.
Copy number alterations are associated with the signature scores
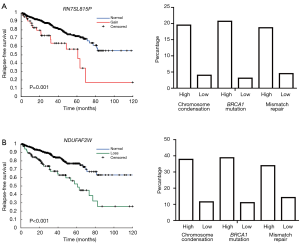
To identify survival associated CNAs, copy number status of each gene was applied to Cox regression model. A total of 198 genes with copy number gains and 337 genes with copy number losses were identified with survival association. Then samples were categorized into two groups based on scores of each gene signature, and chi-square test was used to estimate the proportional difference of copy number changes between the two groups. For the 198 prognostic copy number gains, 197 genes showed significant proportional differences in all three gene sets (P<0.001). RNA5SP73 was the only gene with a proportional difference in the “BRCA1 mutation” signature, but with no statistical difference in the other two. The top 5 prognostic genes are listed in the Table 2. Copy number gains of RN7SL815P was associated with worse survival. We also discovered the proportion of samples with copy number gains in the high signature score groups are higher than those in the low score groups. Comparing with 20% of samples have copy number gains in the high signature score groups, <5% of samples in the low score groups have copy number gains (Figure 3A). For the top 5 genes, RN7SL815P and RN7SL159P are pseudogenes affiliated with the antisense RNA class, and LINC00963 is a long non-coding gene. GAP43 is considered as a neuron growth-associated protein. XPO6 is a member of the importin-beta family. For the 5 genes, the proportions of samples with copy number gains were all above 15% in groups of samples with high scores. On the other hand, the proportions were below 9% in the low-score groups. For the 337 genes with prognostic copy number losses, a total of 185, 273, 156 genes were identified with proportional differences (P<0.001) in the three gene signatures, respectively. The significant percentage in copy number losses is much lower than copy number gain. For the top 5 prognostic genes with copy number losses, the proportion of sample with copy number loss are more than 37% in the groups of high score and only around 10% in the low score groups. Taking NDAUFAF as an example, the percentage of sample with copy number losses was 37.8%, 38.8%, and 33.9% in the high score groups for “chromosome condensation”, “mismatch repair” and “BRCA1 mutation”, respectively. The percentage in low score groups was only 11.5%, 11%, and 14.2%, respectively (Figure 3B).
Table 2
| Gene | HR | 95% CI | p value | Chromosome condensation | BRCA1 mutation | Mismatch repair | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | P value | High | Low | P value | High | Low | P value | ||||||
| Gain | ||||||||||||||
| RN7SL815P | 2.93 | (1.84–4.67) | 5.49E-06 | 38/195 (19.5%) | 9/195 (4.6%) | 9.30E-06 | 40/193 (20.7%) | 7/226 (3.1%) | 3.00E-07 | 37/198 (18.7%) | 10/221 (4.5%) | 4.32E-05 | ||
| RN7SL159P | 2.52 | (1.60–3.97) | 6.17E-05 | 42/189 (22.2%) | 14/189 (7.4%) | 4.85E-05 | 46/182 (25.3%) | 10/233 (4.3%) | 4.93E-08 | 38/195 (19.5%) | 18/220 (8.2%) | 4.15E-03 | ||
| LINC00963 | 2.52 | (1.60–3.96) | 6.36E-05 | 43/189 (22.8%) | 14/189 (7.4%) | 2.94E-05 | 47/182 (25.8%) | 10/233 (4.3%) | 2.76E-08 | 39/195 (20.0%) | 18/220 (8.2%) | 2.83E-03 | ||
| GAP43 | 2.72 | (1.66–4.45) | 7.28E-05 | 37/196 (18.9%) | 9/196 (4.6%) | 1.62E-05 | 39/194 (20.1%) | 7/225 (3.1%) | 5.55E-07 | 36/199 (18.1%) | 10/220 (4.5%) | 7.41E-05 | ||
| XPO6 | 2.56 | (1.61–4.09) | 7.75E-05 | 30/194 (15.5%) | 9/194 (4.6%) | 3.28E-04 | 33/193 (17.1%) | 6/225 (2.7%) | 2.84E-06 | 27/202 (13.4%) | 12/216 (5.6%) | 1.81E-02 | ||
| Loss | ||||||||||||||
| NDUFAF2 | 0.52 | (0.39–0.70) | 8.03E-06 | 65/172 (37.8%) | 25/172 (14.5%) | 2.10E-06 | 66/170 (38.8%) | 24/219 (11.0%) | 3.55E-07 | 60/177 (33.9%) | 30/212 (14.2%) | 4.08E-04 | ||
| SMIM15 | 0.52 | (0.39–0.70) | 8.03E-06 | 65/172 (37.8%) | 25/172 (14.5%) | 2.10E-06 | 66/170 (38.8%) | 24/219 (11.0%) | 3.55E-07 | 60/177 (33.9%) | 30/212 (14.2%) | 4.08E-04 | ||
| KIF2A | 0.53 | (0.40–0.71) | 1.30E-05 | 66/170 (38.8%) | 23/170 (13.5%) | 1.71E-07 | 66/169 (39.1%) | 23/220 (10.5%) | 1.53E-07 | 59/177 (33.3%) | 30/212 (14.2%) | 4.14E-04 | ||
| IPO11 | 0.53 | (0.40–0.71) | 1.30E-05 | 66/170 (38.8%) | 23/170 (13.5%) | 1.71E-07 | 66/169 (39.1%) | 23/220 (10.5%) | 1.53E-07 | 59/177 (33.3%) | 30/212 (14.2%) | 4.14E-04 | ||
| CKS1B | 0.54 | (0.41–0.72) | 2.32E-05 | 65/171 (38.0%) | 22/171 (12.9%) | 1.31E-07 | 65/170 (38.2%) | 22/221 (10.0%) | 1.17E-07 | 57/179 (31.8%) | 30/212 (14.2%) | 8.96E-04 | ||
Discussion
The usage of PARP inhibitors to treat tumors harboring DNA repair defects, especially with homologous recombination deficiency (HRD), is one of the earliest successes of precision oncology (15,16). In prostate cancer, germline and/or somatic mutations of DNA repair genes are closely associated with patients’ response to not only PARP inhibitor (6,17), but also chemotherapy (18,19) and androgen deprivation therapies (20,21). The emerging technology of liquid biopsies, such as circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA), enables a least-invasive assay for these mutations and promises wide clinical applications (22). However, the role of genomic mechanisms other than mutations of the DNA repair genes remains an uncharted territory. In fact, integrative analysis of genomic and proteomic profiles of prostate tumors is realized using next-generation sequencing and RPPA. For instance, integrations of DNA- and RNA-Seq revealed novel classification beyond Gleason score (23) and clinically actionable alterations (24) of prostate cancer. However, such analysis has not been focused on DNA repair genes that can bring biological insights to the observed clinical significance and reveal novel experiment and therapeutic targets. Here, we re-explored the multi-omics profiles of TCGA and carried out a comprehensive analysis of gene expression, functional proteomics, and CNAs of DNA repair genes. Our analysis reached several major findings. First, we conducted a gene set analysis of expression profiles of DNA repair genes. Such analyses have been shown to achieve higher interpretability and tolerance to data noise than investigations at the single-gene level (25-27). As a result, we demonstrated that activities of DNA repair-related functions, including chromosome condensation, mismatch repair, and signaling of BRCA1 mutation, were predictive of progression-free survival of prostate cancer. Our data are in line with previous studies showing that activations of a cell cycle marker and a cell proliferation signature were predictive of adverse survival of prostate cancer (28,29). Second, we demonstrated a concordant prognostic value in protein activities of these genes. Such concordance indicates a meaningful effect of expressional changes in DNA repair genes to really perturb protein functions and affect survival outcomes. Our data underlined many proteins for further investigations. For instance, CDC20 was the top survival predictor among BRCA1 mutation-regulated genes. It is a regulatory protein functioning at several points in the cell division cycle. High expression of CDC20 is associated with high Gleason score, biochemical recurrence, tumor cell growth, and chemoresistance of prostate cancer (30,31). Our data confirmed the unfavorable role of its protein activity. Among proteins associated with chromosome condensation, several non-SMC condensin complexes (NCAPD2, NCAPD3, NCAPH, NCAPH2, NCAPG, and NCAPG2) were associated with adverse survival. These complexes are players in chromosome assembly and segregation, and may be crucial in tumor cell proliferation and tumor progression. Our data warrant further investigations on these genes. At last, CNAs in some DNA repair genes, including two previously unexplored genes NDUFAF2 and RN7SL815P, were indicative of poor survival. NDUFAF2 encodes a mitochondrial protein that is moderately to strongly expressed among cancers according to the Human Protein Atlas (32,33). Our data were the first to demonstrate the prognostic effect of its CNA in prostate cancer. Altogether, the study presents a unique extension from DNA mutations to expressional functions, proteomic activities, and copy numbers of DNA repair genes in prostate cancer. Our findings revealed crucial prognostic markers and candidates for further biological and clinical investigations.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.07.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was waived. This study used public available data which do not require ethics approval.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mucci LA, Hjelmborg JB, Harris JR, et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA 2016;315:68-76. [Crossref] [PubMed]
- Haraldsdottir S, Hampel H, Wei L, et al. Prostate cancer incidence in males with Lynch syndrome. Genet Med 2014;16:553-7. [Crossref] [PubMed]
- Kote-Jarai Z, Leongamornlert D, Saunders E, et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer 2011;105:1230-4. [Crossref] [PubMed]
- Leongamornlert D, Mahmud N, Tymrakiewicz M, et al. Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer 2012;106:1697-701. [Crossref] [PubMed]
- Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016;375:443-53. [Crossref] [PubMed]
- Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 2015;373:1697-708. [Crossref] [PubMed]
- Pomerantz MM, Spisak S, Jia L, et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer 2017;123:3532-9. [Crossref] [PubMed]
- Tian L, Greenberg SA, Kong SW, et al. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci U S A 2005;102:13544-9. [Crossref] [PubMed]
- Li J, Lu Y, Akbani R, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods 2013;10:1046-7. [Crossref] [PubMed]
- Liu W, Tanasa B, Tyurina OV, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010;466:508-12. [Crossref] [PubMed]
- Chen T, Sun Y, Ji P, et al. Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. Oncogene 2015;34:4019-31. [Crossref] [PubMed]
- Zhou Z, He M, Shah AA, et al. Insights into APC/C: from cellular function to diseases and therapeutics. Cell Div 2016;11:9. [Crossref] [PubMed]
- Bunting SF, Callen E, Kozak ML, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 2012;46:125-35. [Crossref] [PubMed]
- Nicolas E, Golemis EA, Arora S. POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene 2016;590:128-41. [Crossref] [PubMed]
- Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature 2012;481:287-94. [Crossref] [PubMed]
- La Thangue NB, Kerr DJ. Predictive biomarkers: a paradigm shift towards personalized cancer medicine. Nat Rev Clin Oncol 2011;8:587-96. [Crossref] [PubMed]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244-50. [Crossref] [PubMed]
- Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol 2013;31:1748-57. [Crossref] [PubMed]
- Cheng HH, Salipante SJ, Nelson PS, et al. Polyclonal BRCA2 Reversion Mutations Detected in Circulating Tumor DNA After Platinum Chemotherapy in a Patient With Metastatic Prostate Cancer. JCO Precis Oncol 2018. doi:
10.1200/PO.17.00176 . - Antonarakis ES, Lu C, Luber B, et al. Germline DNA-repair Gene Mutations and Outcomes in Men with Metastatic Castration-resistant Prostate Cancer Receiving First-line Abiraterone and Enzalutamide. Eur Urol 2018;74:218-25. [Crossref] [PubMed]
- Annala M, Struss WJ, Warner EW, et al. Treatment Outcomes and Tumor Loss of Heterozygosity in Germline DNA Repair-deficient Prostate Cancer. Eur Urol 2017;72:34-42. [Crossref] [PubMed]
- Vandekerkhove G, Chi KN, Wyatt AW. Clinical utility of emerging liquid biomarkers in advanced prostate cancer. Cancer Genet 2018;228-229:151-8. [Crossref] [PubMed]
- Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11-22. [Crossref] [PubMed]
- Robinson D, Van Allen EM, Wu YM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015;162:454. [Crossref] [PubMed]
- Lin EP, Hsiao TH, Lu JY, et al. Translating Gene Signatures Into a Pathologic Feature: Tumor Necrosis Predicts Disease Relapse in Operable and Stage I Lung Adenocarcinoma. JCO Precis Oncol 2018. doi:
10.1200/PO.18.00043 . - Hsiao TH, Chiu YC, Hsu PY, et al. Differential network analysis reveals the genome-wide landscape of estrogen receptor modulation in hormonal cancers. Sci Rep 2016;6:23035. [Crossref] [PubMed]
- Hsu YC, Chiu YC, Chen Y, et al. A simple gene set-based method accurately predicts the synergy of drug pairs. BMC Syst Biol 2016;10:66. [Crossref] [PubMed]
- Yang RM, Naitoh J, Murphy M, et al. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol 1998;159:941-5. [Crossref] [PubMed]
- Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12:245-55. [Crossref] [PubMed]
- Li K, Mao Y, Lu L, et al. Silencing of CDC20 suppresses metastatic castration-resistant prostate cancer growth and enhances chemosensitivity to docetaxel. Int J Oncol 2016;49:1679-85. [Crossref] [PubMed]
- Mao Y, Li K, Lu L, et al. Overexpression of Cdc20 in clinically localized prostate cancer: Relation to high Gleason score and biochemical recurrence after laparoscopic radical prostatectomy. Cancer Biomark 2016;16:351-8. [Crossref] [PubMed]
- Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248-50. [Crossref] [PubMed]
- Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]


