Construction and analysis of lncRNA-associated ceRNA network identified potential prognostic biomarker in gastric cancer
Introduction
Gastric cancer (GC) is known as the fifth most common malignancy with nearly 952,000 new cases in 2012, mostly occurs in eastern Asia. It is the third leading cause of cancer death worldwide, comprising 8.8% of total cancer deaths (1). The five-year survival rate of GC stage I and IIA is around 81.8–93.6%, meanwhile that of the stage IIIC is 17.9% (2). Except for radical resection, the therapy options for advanced GC are limited. Thus, searching for the sensitive biomarkers to detect GC in early stage and treatment targets for effective individualized therapy is urgent and crucial.
In total, 75% of the human genome is transcribed to RNA, but 90% of RNAs are without protein-coding function, defined as non-coding RNAs (ncRNAs), which were considered as “transcriptional noise” previously (3). ncRNAs consist of small ncRNAs and long non-coding RNAs (lncRNAs), which are transcripts longer than 200 nucleotides with tissue specificity (4). In these decades, lncRNAs were found playing an important role in gene regulations (5-7). In 2011, Salmena et al. put forward the competitive endogenous RNA (ceRNA) hypothesis to describe a complex post-transcriptional regulatory network, with components of lncRNAs, mRNAs, other RNAs (circular RNA and pseudogene) and microRNA (miRNA). The RNA-RNA crosstalk is working with the principle, that lncRNAs and other RNAs compete the target RNA by sponging miRNA, the common miRNA response elements (MREs) (8). Increasing evidences demonstrated that the lncRNAs were involved in tumorigenesis and progression of breast cancer, liver cancer, lung adenocarcinoma, colorectal cancer (CRC), ovarian cancer and other malignancies (9-13)
Recent studies have revealed several lncRNAs that differentially expressed in GC, as well as their corresponding mechanisms. HOTAIR, the noted lncRNA, modulates HER2 via competing with miR-331-3p in GC (11,14,15). Previous studies indicates that lncRNAs such like GAPLINC, FER1L4, H19 and GACAT1 may interactions with the well-known onco-mRNAs, as CD44 mRNA, RB1, c-Myc and p53, via sponging the common MREs in GC (16-18). However, a whole map of GC specific ceRNA regulatory network and underlying interactions, with large-scale samples need further exploration.
In our study, 375 GC samples and 32 normal samples were obtained from The Cancer Genome Atlas (TCGA) Stomach Adenocarcinoma (STAD) datasets to detect the differentially expression of lncRNAs, miRNAs, and mRNAs. Statistics were further improved the reliability and accuracy by matching the RNA-RNA interaction pairs. The lncRNA associated ceRNA network was constructed, and the function and prognostic value of the key lncRNAs were analyzed.
Methods
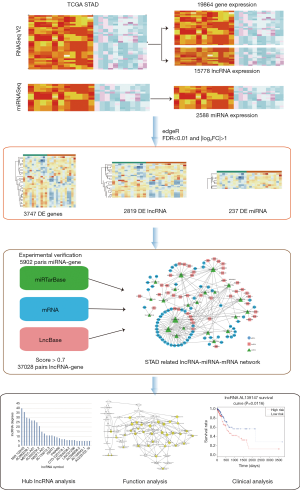
The steps of the statistics analyzing were shown in Figure 1.
Date collection
The RNAseq and lncRNAseq of 375 GC samples and 32 normal samples were obtained from TCGA STAD datasets with gtf file of GENCODE version 22. Meanwhile, miRNAseq isoform counts data of 446 GC samples and 45 normal samples were downloaded from the TCGA. To standardize the statistics of the miRNAseq, the 2,588 miRNA mature IDs from miRBase were cross matched with the above data. Then miRNAs were deleted if negative expression in up to 80% samples. We enrolled the 404 samples with date of both the RNAseq and miRNAseq, 365 of which were clinical follow-up. Our research followed the publication guidelines provided by TCGA Network (http://cancergenome.nih.gov/publications/publicationguidelines).
Differentially expressed analysis
We identify differentially expressed (DE) mRNAs, miRNA and lncRNAs by applying edge R package with thresholds of |log2FC| >2 and false discovery rate (FDR) <0.01 [fold change (FC)].
Construction of the ceRNA network
To predict target genes, we downloaded the miRNA-mRNA interactions with low throughput experimentally verified from miTarbase database. We defined the pairs in the LncBase database, which contains crosslinking immunoprecipitation sequencing (CLIP-Seq) verified and computationally predicted miRNA-lncRNA interactions, with scores greater than 0.7 as the potential ones.
Further, we calculated the Pearson’s correlation coefficient of DEmiRNAs and DEmRNAs, that of DElncRNAs and DEmRNAs as well as that of DElncRNAs and DEmiRNAs respectively. According to the ceRNA hypothesis, we draw criteria for the candidates of ceRNA network as following: (I) mRNAs and lncRNAs share the same miRNAs. (II) Positive correlation showed between mRNAs and lncRNAs using Pearson’s correlation coefficient (r>0.3, P<0.05). Negative correlation should be investigated between mRNAs and lncRNAs, between mRNAs and miRNAs as well as between lncRNAs and miRNAs (r<−0.3, P<0.05). According to the above potential interactions of miRNA-mRNA-lncRNA, the ceRNA network was constructed with Cytoscape.
Network analysis and Functional Enrichment analysis
The constructed ceRNA network was analyzed the topological properties: the degree, betweenness and closeness of the nodes in the net were calculated with the built-in Network analyzer tool in the Cytoscape.
With collecting the top 5 degree of lncRNAs in ceRNA network, we perform the Gene oncology (GO) function enrichment in BINGO plugin of Cytoscape to detect the potential biological function.
Survival analysis
To identify the prognostic lncRNAs, we inspect the differentiate expression RNAs combined with the clinical data from the 365 GC samples in TCGA. Kaplan-Meier survival analysis was applied to analyze the correlation between lncRNA expression and clinical prognosis.
Results
Differentiate expression of lncRNAs, mRNAs and miRNAs in GC
A total of 19,836 mRNAs, 15,778 lncRNAs and 2,588 miRNAs were identified out from 404 samples in TCGA. With thresholds of |log2FC| >2 and FDR <0.01 in edge R package, a total of 3,747 mRNAs (1,719 up-regulated and 2,028 down-regulated), 2,819 lncRNAs (1,909 up-regulated and 910 down-regulated) and 237 miRNAs (156 up-regulated and 81 down-regulated) were detected differentially expressed in GC tissues compared to the control ones. The DEmRNAs, DEmiRNAs and DElncRNAs with top 100 FDR value were shown on the differentiate expression heatmaps (Figure S1A,B,C).
Construction of the ceRNA network
To detect the reliable interactions, 380,257 miRNA-mRNA interactions with low throughput experimentally verified in miTarbase database were chosen to compare with the DEmiRNAs and DEmRNAs data. In total, 5,902 pairs of miRNA-mRNA were met with both conditions. To improve the predictive accuracy, 2,573,729 miRNA-lncRNA pairs, which we selected from the LncBase database with the threshold set to 0.7, were matched with DEmiRNAs and DElncRNAs data. Thus, 37,028 miRNA-lncRNA pairs were identified. According to the ceRNA hypothesis, 443 mRNA-lncRNA pairs who shared with the same miRNA were involved. Then we further collected 431 mRNA-lncRNA pairs with positive correlationship (Pearson’s correlation analysis, r>0.3, P<0.01), 102 lncRNA-miRNA pairs with negative correlationship (r<−0.3, P<0.01) and 109 mRNA-miRNA with negative correlationship (r<−0.3, P<0.01) as the candidates for ceRNA network.
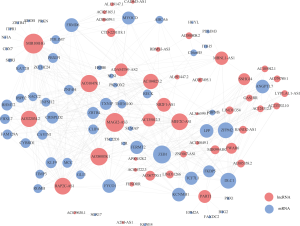
The ceRNA network involved 61 mRNAs, 44 lncRNAs and 22 miRNAs was visualized using Cytoscape software based on the 211 interactions among them (Figure 2). 181 interactions between lncRNAs and mRNAs were presented in Figure 3.
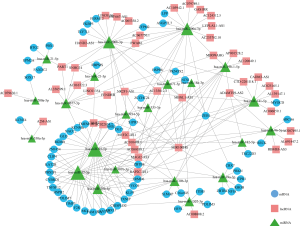
Network analysis and Functional Enrichment analysis of DElncRNAs
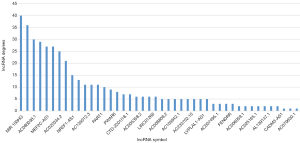
To analyze the topological properties of the ceRNA network, the degree, closeness and betweenness of RNAs were calculated. The degree distribution of the DEmiRNAs was shown in Table 1. The top 5 highest degree in network were hsa-miR-93-5p, hsa-miR-17-5p, hsa-miR-200a-3p, hsa-miR-200b-3p and hsa-miR-141-5p. The degree distribution of the DElncRNAs in lncRNA-mRNA network (Table 2) with their corresponding competitive mRNAs was displayed in Figure 4, and the top 5 highest degree of DElncRNAs were MIR100HWG, MAGI2-AS3, AC080038.1, AC010478.1 and MEF2C-AS1. The competitive mRNAs of lncRNAs shown in Figure 4 are related to tumorigenesis, as ZEB1, BTG2, MYOCD and SRP (19-21).
Table 1
| miRNA | Degree |
|---|---|
| hsa-miR-93-5p | 29 |
| hsa-miR-17-5p | 27 |
| hsa-miR-200a-3p | 19 |
| hsa-miR-200b-3p | 18 |
| hsa-miR-141-5p | 13 |
| hsa-miR-19b-1-5p | 11 |
| hsa-miR-20a-5p | 11 |
| hsa-miR-130b-3p | 9 |
| hsa-miR-19a-3p | 9 |
| hsa-miR-4677-3p | 9 |
| hsa-miR-96-5p | 9 |
| hsa-miR-1307-3p | 7 |
| hsa-miR-671-5p | 7 |
| hsa-miR-182-5p | 5 |
| hsa-miR-21-3p | 5 |
| hsa-miR-21-5p | 5 |
| hsa-miR-335-3p | 5 |
| hsa-miR-3934-5p | 4 |
| hsa-miR-18a-3p | 3 |
| hsa-miR-200a-5p | 2 |
| hsa-miR-301b-3p | 2 |
| hsa-miR-550a-3p | 2 |
DEmiRNA, differentially expressed miRNA; ceRNA, competitive endogenous RNA.
Table 2
| lncRNA | Degree |
|---|---|
| MIR100HG | 40 |
| MAGI2-AS3 | 36 |
| AC080038.1 | 30 |
| AC010478.1 | 29 |
| MEF2C-AS1 | 27 |
| RAP2C-AS1 | 27 |
| AC022034.2 | 25 |
| AC104825.2 | 21 |
| NR2F1-AS1 | 15 |
| MBNL1-AS1 | 13 |
| AC135012.3 | 11 |
| ADAMTS9-AS2 | 11 |
| PART1 | 11 |
| SNHG14 | 10 |
| PWAR6 | 9 |
| AC067750.1 | 8 |
| CTD-2201I18.1 | 7 |
| MIR99AHG | 7 |
| AC005358.2 | 6 |
| HAND2-AS1 | 6 |
| LINC01266 | 6 |
| LINC01354 | 6 |
| AC008808.2 | 5 |
| AC008808.2 | 5 |
| AC079789.1 | 5 |
| AC105942.1 | 5 |
| AC124312.5 | 5 |
| AC233702.10 | 5 |
| GAS1RR | 5 |
| LYPLAL1-AS1 | 5 |
| ZNF667-AS1 | 5 |
| AC007495.1 | 3 |
| AL691447.2 | 3 |
| FENDRR | 3 |
| RBMS3-AS3 | 3 |
| AC006059.1 | 2 |
| AC016722.3 | 2 |
| AC025165.1 | 2 |
| AC120049.1 | 2 |
| AL139147.1 | 2 |
| AP001528.2 | 2 |
| CADM3-AS1 | 2 |
| A2M-AS1 | 1 |
| AC079630.1 | 1 |
| AL356599.1 | 1 |
lncRNA, long non-coding RNA; ceRNA, competitive endogenous RNA.
GO enrichment analyses were performed in BINGO plugin of Cytoscape to detect the potential biological functions of the top 5 highest degree of DElncRNAs. The 5 lncRNAs target genes except AC010478.1 were significantly enriched in GO biological process categories, mostly tumor-related functions, as “cellular metabolic process”, “cell proliferation” and “canonical Wnt receptor signaling pathway” (P<0.05) (Figure S2). The function enrichment of the lncRNA AC080038.1 target genes in ceRNA network were demonstrated in Figure 5.
Survival analysis
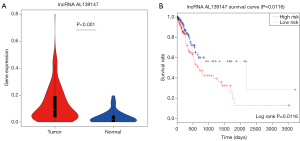
According to analyzed the clinical characteristics and outcome, the lncRNA AL139147 were identified significantly affecting prognosis (HR 5.884, P<0.05) (Table S1). AL139147 differentially expressed in GC samples (log2FC =1.407, P<0.001) (Figure 6A). Kaplan-Meier analysis was applied to investigate overall survival for AL139147 in GC samples. As a result, AL139147 were detected negatively correlated with overall survival by univariate Cox regression analysis (log-rank, P=0.0116) (Figure 6B).
Discussion
Decades of evidences have demonstrated that the carcinogenesis and progress of cancer is a complex multiple “hits” process with breaking the balance between oncogene and tumor suppressor gene, rather than up or down regulation of one simple gene. The recent ceRNA hypothesis accord with it, which explains the lncRNAs function as the post-transcriptional gene regulator. LncRNAs were involved in the ceRNA network, cross talking with specific RNAs (including mRNAs, circular RNAs and pseudogenes) via sponging the MREs and upregulating the target RNA expression subsequently (7,8). The imbalance of the ceRNA network may lead to various diseases. The ceRNA network has been constructed and further proved in breast cancer, liver cancer, lung adenocarcinoma, CRC, ovarian cancer and other malignancies, helping us discover crucial lncRNAs as the new diagnostic or therapeutic targets (9,10,12,13,22). Several lncRNAs have been detected playing an important role as members in ceRNA network in GC and closely related to prognosis. HOTAIR was a better-characterized lncRNAs in GC as ceRNAs, modulating the depression of HER2, which was related to lymph node metastasis and poor prognosis, via competing with miR-331-3p (11,14). The lncRNA GAPLINC functions as the miRNA 211-3p sponge to regulate CD44 mRNA, which involved in tumor proliferation, angiogenesis and migration (16,17). lncRNAs such as FER1L4, H19, TUSC7 and GACAT1 may interact with the well-known onco-mRNA, like RB1, PTEN, CDKN1A, VEGFA, E2F1, IL-10, HIPK3, PAK7 or P 53 in GC through preliminary studies (18,23).
In present study, 44 lncRNAs were detected to participate in the constructed ceRNA network, with differential expression and target miRNA as well as correlationship of interactions screening to obtain high reliable algorithms. MIR100HG, MAGI2-AS3, AC080038.1, AC010478.1 and MEF2C-AS1 ranked as the most involved lncRNAs in this ceRNA network by topological features. Their target genes were enriched in GO biological process categories as “cellular metabolic process”, “cell proliferation” and “regulation of Wnt receptor signaling pathway”, which were closely related to tumorigenesis and progression. The lncRNA MIR100HG, located in chromosome 11, is a miRNA host gene which encodes miR-100, let-7a-2 and miR-125-b. It was reported holding an intronic coding region, which makes it as a proapoptotic molecule through a caspase-dependent mitochondrial pathway of cell death (24). It is also correlated with the gap junction and TGF-β pathways to participate the tumor development (25). The high expression of MIR100HG is detected in acute megakaryoblastic leukemia as well as cervical cancer, and is associated with the poor prognosis. Analysis of TCGA CRC date found that increase expression of MIR100HG with stage-dependent pattern. Researchers applied MIR100HG as the diagnostic method for cervical cancer, and the ROC curve showed promising discrimination power with areas under the curve of 0.801 (26). Furthermore, Lu et al. revealed that MIR100HG mediate de novo and acquired cetuximab-resistant in CRC cells via Wnt signaling (27). Overexpression of miR-100 and miR-125-b has been observed in GC, but how the expression of MIR100HG changed in GC and the way it involved as the ceRNA, need further research. MAGI2-AS3 is a newly discovered lncRNA, recognized as a tumor suppressor in breast cancer, via regulating Fas/FasL signaling pathway. Breast cancer cells low expressed MAGI2-AS3. And MAGI2-AS3 expression was negative related with histological grade, TNM stage and HER-2 expression (28). lncRNA MEF2C-AS1 is an antisense RNA to MEF2C, which is identified as the potential oncogene in T cell acute lymphoblastic leukemia (29). Study revealed that MEF2C might be the molecular mechanism of chemotherapy resistance in acute myeloid leukemia (30). Luo et al. showed that lncRNA MEF2C-AS1 significantly downregulated in diffuse GC, and the in-vitro assays detected that the knock-down of MEF2C-AS1 promoted aggressive tumor behaviors (31). Another highlight lncRNA, AL139147 is located in chromosome 1 and is suspected with disease as psoriasis, atrioventricular septal defect and peripheral arterial occlusive till now. It regulated the mRNA SGIP1 and DKFZp761D221 variant protein. Although the connection between AL139147 and cancer has not been found yet, the expression of AL139147 in GC samples was significantly higher than that in normal ones through analysis of TCGA database. Meanwhile, our constructed network indicated that it interplayed with other ceRNAs and miRNAs in the GC as a network node, targeting MYOCD and FRMD6, which were contributing to tumor development pathway (32). Furthermore, according to the Kaplan Meier analysis of the clinical statistics of the 365 samples, high expression of AL139147 has a tendency of poor prognosis (P<0.05). Till now, there has been few research on lncRNA AL139147, AC080038.1 as well as AC01047. Evidences suggest that our constructed ceRNA network is highly associated with tumorigenesis and chemotherapy resistance, and the crucial lncRNAs in the network might be promising biomarker and therapeutic target for GC.
To draw a comprehensive of the lncRNA-miRNA-mRNA crosstalk in the network, we listed the top five highest degree miRNA, which were hsa-miR-93-5p, hsa-miR-17-5p, hsa-miR-200a-3p, hsa-miR-200b-3p and hsa-miR-141-5p. According to literature review, hsa-miR-93-5p, hsa-miR-17-5p, and miR-141-5p were significantly up-regulated in 75.8% CRC, breast cancer and ovarian cancer respectively (33-35). A negative correlation of hsa-miR-17-5p with overall survival was found in TCGA breast cancer specimens, and miRNA-200 family members were detected associated with brain metastases of gastric adenocarcinoma (36). The lncRNA target genes, as ZEB1, BTG2, MYOCD and FRMD6 are strongly related to tumorigenesis and progression. ZEB1 is widely known as the activator of an epithelial-mesenchymal transition (EMT) and has important roles in metastasis. Mounting evidences showed the double-negative feedback loop of miR-200 and ZEB1 control multiple genes involved in migration, invasion and metastasis (21). Furthermore, it might play a role in tumor regulation of PD-L1, leading to intratumoral immunosuppression (37). MYOCD, contacting with SRF, functions in cardiac muscle and smooth muscle, plays a role in nasopharyngeal carcinoma, breast cancer, and uterine leiomyosarcoma (38,39). BTG2 is known as the tumor suppressor, take part in cell differentiation, proliferation, DNA damage repair and apoptosis (40). The down regulation of BTG2 was detected in breast cancer, renal clear cell carcinoma, GC and prostate cancer. In studies, it was related to tumor size, grade, metastasis, recurrence and poor survival in patients with breast cancer (41). FRMD6 expression activates the Hippo signaling pathway regulating organ size control, cell proliferation and cancer development and antagonizes oncogenic YAP (32). Clinically, it has been identified from microarray data as significantly predictor for the survival of patients with CRC compared to the conventional Dukes’ classification (42). In our ceRNA network, the above genes interact with multiple lncRNAs, which indicates that the lncRNAs in the network are worthy of our researching.
In conclusion, our study constructed a high confidence level of ceRNA network by genome-wide analysis of gastric adenocarcinoma patients from TCGA database with strict filtering criteria according to lncRNA-mRNA-miRNA inner logical relationships. The dysregulated of the cancer specific lncRNAs namely MIR100HG, MAGI2-AS3, AC080038.1, AC010478.1 and MEF2C-AS1 were identified as the candidate for diagnostic biomarker and therapeutic targets. lncRNA AL139147 might be a promising biomarkers for predicting prognosis. Our research may help us further understanding of the ceRNA pathogenesis in GC and guide for further investigation.

Table S1
| lncRNA | ENSG | HR | P value | Z |
|---|---|---|---|---|
| MIR100HG | ENSG00000255248 | 1.048411 | 0.175386 | 1.355098 |
| MAGI2-AS3 | ENSG00000234456 | 1.242736 | 0.06959 | 1.81457 |
| AC080038.1 | ENSG00000274565 | 3.360686 | 0.065983 | 1.838537 |
| AC010478.1 | ENSG00000259663 | 1.182393 | 0.385268 | 0.86823 |
| MEF2C-AS1 | ENSG00000248309 | 1.749401 | 0.552753 | 0.59364 |
| RAP2C-AS1 | ENSG00000232160 | 1.050688 | 0.822708 | 0.224064 |
| AC022034.2 | ENSG00000237807 | 0.969829 | 0.614905 | −0.50308 |
| AC104825.2 | ENSG00000251615 | 1.056401 | 0.573507 | 0.562894 |
| NR2F1-AS1 | ENSG00000237187 | 1.288318 | 0.159086 | 1.408151 |
| MBNL1-AS1 | ENSG00000229619 | 0.991053 | 0.737805 | −0.33476 |
| AC135012.3 | ENSG00000268505 | 1.245253 | 0.629784 | 0.482031 |
| ADAMTS9-AS2 | ENSG00000241684 | 1.042312 | 0.899259 | 0.126598 |
| PART1 | ENSG00000152931 | 1.063839 | 0.620626 | 0.494964 |
| SNHG14 | ENSG00000224078 | 1.166315 | 0.407183 | 0.82886 |
| PWAR6 | ENSG00000257151 | 0.997848 | 0.989369 | −0.01332 |
| AC067750.1 | ENSG00000272631 | 1.047768 | 0.901524 | 0.123736 |
| CTD-2201I18.1 | ENSG00000249825 | 1.319286 | 0.336403 | 0.961297 |
| MIR99AHG | ENSG00000215386 | 1.617249 | 0.037948 | 2.075419 |
| AC005358.2 | ENSG00000265489 | 6.525081 | 0.257969 | 1.131205 |
| HAND2-AS1 | ENSG00000237125 | 0.984551 | 0.605558 | −0.51642 |
| LINC01266 | ENSG00000224957 | 3.270522 | 0.342831 | 0.948586 |
| LINC01354 | ENSG00000231768 | 2.658016 | 0.228987 | 1.202973 |
| AC008808.2 | ENSG00000250360 | 1.040367 | 0.728891 | 0.346601 |
| AC079789.1 | ENSG00000261672 | 1.566297 | 0.392122 | 0.855776 |
| AC105942.1 | ENSG00000235501 | 1.021449 | 0.650374 | 0.453242 |
| AC124312.5 | ENSG00000271347 | 1.0101 | 0.948257 | 0.064896 |
| AC233702.10 | ENSG00000272780 | 3289.664 | 0.287533 | 1.06355 |
| GAS1RR | ENSG00000226237 | 2.448866 | 0.227506 | 1.206807 |
| LYPLAL1-AS1 | ENSG00000228063 | 0.715678 | 0.774458 | −0.28655 |
| ZNF667-AS1 | ENSG00000166770 | 1.038709 | 0.601017 | 0.522939 |
| AC007495.1 | ENSG00000246379 | 1.699957 | 0.577916 | 0.556431 |
| AL691447.2 | ENSG00000232063 | 1.542672 | 0.40415 | 0.834232 |
| FENDRR | ENSG00000268388 | 1.011148 | 0.650783 | 0.452674 |
| RBMS3-AS3 | ENSG00000235904 | 1.083634 | 0.864213 | 0.171014 |
| AC006059.1 | ENSG00000230084 | 1.419768 | 0.335101 | 0.963891 |
| AC016722.3 | ENSG00000260977 | 1.097406 | 0.845642 | 0.194682 |
| AC025165.1 | ENSG00000224713 | 1.232356 | 0.676442 | 0.417323 |
| AC120049.1 | ENSG00000267414 | 1.392001 | 0.242593 | 1.16853 |
| AL139147.1 | ENSG00000248458 | 5.839658 | 0.016491 | 2.397861 |
| AP001528.2 | ENSG00000255471 | 1.547054 | 0.05401 | 1.926756 |
| CADM3-AS1 | ENSG00000225670 | 2.802363 | 0.064674 | 1.847502 |
| A2M-AS1 | ENSG00000245105 | 1.011894 | 0.929037 | 0.089057 |
| AC079630.1 | ENSG00000225342 | 2.810959 | 0.173474 | 1.361126 |
| AL356599.1 | ENSG00000235652 | 0.679064 | 0.504824 | −0.66692 |
lncRNA, long non-coding RNA.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.06.32). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our research followed the publication guidelines provided by TCGA Network (http://cancergenome.nih.gov/publications/publicationguidelines). The institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Amin MB, Greene FL, Edge SB, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin 2017;67:93-9.
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013;152:1308-23. [Crossref] [PubMed]
- Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014;9:3-12. [Crossref] [PubMed]
- Orom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010;143:46-58. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. [Crossref] [PubMed]
- Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83. [Crossref] [PubMed]
- Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014;13:92. [Crossref] [PubMed]
- Hou P, Zhao Y, Li Z, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 2014;5:e1287. [Crossref] [PubMed]
- Sui J, Li YH, Zhang YQ, et al. Integrated analysis of long non-coding RNA‑associated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol 2016;49:2023-36. [Crossref] [PubMed]
- Lemoine NR, Jain S, Silvestre F, et al. Amplification and overexpression of the EGF receptor and c-erbB-2 proto-oncogenes in human stomach cancer. Br J Cancer 1991;64:79-83. [Crossref] [PubMed]
- Leung TW, Cheung AN, Cheng DK, et al. Expressions of c-erbB-2, epidermal growth factor receptor and pan-ras proto-oncogenes in adenocarcinoma of the cervix: correlation with clinical prognosis. Oncol Rep 2001;8:1159-64. [PubMed]
- Denzler R, Agarwal V, Stefano J, et al. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 2014;54:766-76. [Crossref] [PubMed]
- Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet 2015;16:113-26. [Crossref] [PubMed]
- Gu Y, Chen T, Li G, et al. LncRNAs: emerging biomarkers in gastric cancer. Future Oncol 2015;11:2427-41. [Crossref] [PubMed]
- Zhang L, Jambusaria A, Hong Z, et al. SOX17 Regulates Conversion of Human Fibroblasts Into Endothelial Cells and Erythroblasts by Dedifferentiation Into CD34(+) Progenitor Cells. Circulation 2017;135:2505-23. [Crossref] [PubMed]
- Mao B, Zhang Z, Wang G. BTG2: a rising star of tumor suppressors Int J Oncol 2015;46:459-64. (review). [Crossref] [PubMed]
- Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev 2009;28:151-66. [Crossref] [PubMed]
- Xiong Y, Wang R, Peng L, et al. An integrated lncRNA, microRNA and mRNA signature to improve prognosis prediction of colorectal cancer. Oncotarget 2017;8:85463-78. [Crossref] [PubMed]
- Hu Y, Tian H, Xu J, et al. Roles of competing endogenous RNAs in gastric cancer. Brief Funct Genomics 2016;15:266-73. [Crossref] [PubMed]
- Broustas CG, Gokhale PC, Rahman A, et al. BRCC2, a novel BH3-like domain-containing protein, induces apoptosis in a caspase-dependent manner. J Biol Chem 2004;279:26780-8. [Crossref] [PubMed]
- Noordhuis MG, Fehrmann RS, Wisman GB, et al. Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin Cancer Res 2011;17:1317-30. [Crossref] [PubMed]
- Shang C, Zhu W, Liu T, et al. Characterization of long non-coding RNA expression profiles in lymph node metastasis of early-stage cervical cancer. Oncol Rep 2016;35:3185-97. [Crossref] [PubMed]
- Lu Y, Zhao X, Liu Q, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med 2017;23:1331-41. [Crossref] [PubMed]
- Yang Y, Yang H, Xu M, et al. Long non-coding RNA (lncRNA) MAGI2-AS3 inhibits breast cancer cell growth by targeting the Fas/FasL signalling pathway. Hum Cell 2018;31:232-41. [Crossref] [PubMed]
- Laszlo GS, Alonzo TA, Gudgeon CJ, et al. Erratum to: High expression of myocyte enhancer factor 2C (MEF2C) is associated with adverse-risk features and poor outcome in pediatric acute myeloid leukemia: a report from the Children's Oncology Group. J Hematol Oncol 2016;9:133. [Crossref] [PubMed]
- Brown FC, Still E, Koche RP, et al. MEF2C Phosphorylation Is Required for Chemotherapy Resistance in Acute Myeloid Leukemia. Cancer Discov 2018;8:478-97. [Crossref] [PubMed]
- Luo T, Zhao J, Lu Z, et al. Characterization of long non-coding RNAs and MEF2C-AS1 identified as a novel biomarker in diffuse gastric cancer. Transl Oncol 2018;11:1080-9. [Crossref] [PubMed]
- Angus L, Moleirinho S, Herron L, et al. Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene 2012;31:238-50. [Crossref] [PubMed]
- Paul S, Lakatos P, Hartmann A, et al. Identification of miRNA-mRNA Modules in Colorectal Cancer Using Rough Hypercuboid Based Supervised Clustering. Sci Rep 2017;7:42809. [Crossref] [PubMed]
- Li Z, Peng Z, Gu S, et al. Global Analysis of miRNA-mRNA Interaction Network in Breast Cancer with Brain Metastasis. Anticancer Res 2017;37:4455-68. [PubMed]
- Bartoszewski R, Serocki M, Janaszak-Jasiecka A, et al. miR-200b downregulates Kruppel Like Factor 2 (KLF2) during acute hypoxia in human endothelial cells. Eur J Cell Biol 2017;96:758-66. [Crossref] [PubMed]
- Minn YK, Lee DH, Hyung WJ, et al. MicroRNA-200 family members and ZEB2 are associated with brain metastasis in gastric adenocarcinoma. Int J Oncol 2014;45:2403-10. [Crossref] [PubMed]
- Chen L, Gibbons DL, Goswami S, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 2014;5:5241. [Crossref] [PubMed]
- Malysheva V, Mendoza-Parra MA, Saleem MA, et al. Reconstruction of gene regulatory networks reveals chromatin remodelers and key transcription factors in tumorigenesis. Genome Med 2016;8:57. [Crossref] [PubMed]
- Villacis RA, Silveira SM, Barros-Filho MC, et al. Gene expression profiling in leiomyosarcomas and undifferentiated pleomorphic sarcomas: SRC as a new diagnostic marker. PLoS One 2014;9:e102281. [Crossref] [PubMed]
- Donato LJ, Suh JH, Noy N. Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res 2007;67:609-15. [Crossref] [PubMed]
- Mollerstrom E, Kovacs A, Lovgren K, et al. Up-regulation of cell cycle arrest protein BTG2 correlates with increased overall survival in breast cancer, as detected by immunohistochemistry using tissue microarray. BMC Cancer 2010;10:296. [Crossref] [PubMed]
- Abdul Aziz NA, Mokhtar NM, Harun R, et al. A 19-Gene expression signature as a predictor of survival in colorectal cancer. BMC Med Genomics 2016;9:58. [Crossref] [PubMed]