PDZRN4-mediated colon cancer cell proliferation and dissemination is regulated by miR-221-3p
Introduction
Despite rapid advances in diagnostic and treatment strategies, colon cancer still ranks high among commonly diagnosed cancers, and its incidence is rising (1). In China, the incidence in men and women is 16.9 and 11.6 per 100,000, respectively (2). Colon cancer is closely associated with other diseases such as diabetes and obesity, making it a major public health risk (3,4). Despite the rapid increase in newly diagnosed patients, the etiology and mechanism of colon cancer development remains unclear.
The gene PDZRN4 (PDZ domain containing ring finger 4) belongs to the ligand of numb protein-X (LNX) family and is dysregulated in multiple diseases, including radiation-induced papillary thyroid carcinoma, rectal adenocarcinoma, and hepatocellular carcinoma (5,6). In human liver cancer cell lines, the ectopic expression of PDZRN4 inhibited cancer cell proliferation, as well as plate colony formation and anchorage-independent colony formation (7). Analysis of gene expression data indicates that the gene PDZRN4 is downregulated in colon cancer. In the present study, we investigated the role of PDZRN4 in colon cancer development.
MicroRNAs (miRNAs) are a small noncoding RNA that, by binding to a coding sequence or 3'-untranslated region (UTR), can interfere with mRNA translation or stability, changing expression of the gene. Depending on its downstream gene, miRNAs can function as either tumor suppressors or oncogenes. Because miRNAs have such roles in the development of cancers (8-10), we wondered whether miRNAs may be implicated in the differential expression of PDZRN4 in colon cancer.
Specifically, miR-221 is a putative regulator in tumor development (11-13), and according to open access data in the National Center for Biotechnology Information (NCBI) database its levels are usually elevated in colon cancer (GSE101502). In addition, higher level of miR-221 in colon cancer tissues is associated with poor prognosis (14,15). The expression of miR-221 is regulated by multiple regulators, including KRAS, NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells), STAT3 (signal transducer and activator of transcription 3), and KRAP (Ki-ras-induced actin-interacting protein), while KRAP-regulated miR-221 expression could only be seen in a 3D culture, and NF-κB and STAT3 activation is, in turn, manipulated by miR-221 expression (16,17).
It is possible that miRNAs can have multiple targets, and the link between miR-221 and PDZRN4 remains unclear. To contribute information toward the diagnosis and treatment of colon cancer, the present study explored the potential regulation of PDZRN4 by miR-221-3p.
Methods
Ethics statement
The Institutional Ethics Committee of Second Affiliated Hospital of Soochow University (Suzhou, 215004, Jiangsu Province, China) reviewed and authorized the clinical section of this research.
Colon cancer tissue samples
Twenty pairs of colon cancer tissues and adjacent normal tissues were collected from patients who received treatment in our hospital. None of the patients received radiotherapy or chemotherapy before surgery (Table S1). All diagnoses were obtained via pathological examination. All examinations and evaluations of clinical samples were conducted using paired normal tissue as a control.
Cell line
Cells of the human colon cancer cell line, HCT116 were purchased from the American Type Culture Collection and cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) at 37 °C, 5% CO2.
293T cells were previously preserved in our lab and cultured in Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY, USA) containing 10% FBS and 2 mM L-glutamine at 37 °C, 5% CO2.
Cells were observed with a microscope (Olympus, Tokyo, Japan) and green fluorescent protein (GFP) was observed with a fluorescence microscope.
Plasmids
The pLVX-PDZRN4 plasmid was constructed by inserting the PDZRN4 coding sequence into the lentiviral expression vector, pLVX-IRES-ZsGreen1. The pLVX-miR-221-3p sponge was constructed by referring to a published article (18).
Immunohistochemistry (IHC)
The cancer tissues and paired adjacent normal tissues obtained from patients during surgery were processed for staining with hematoxylin and eosin and IHC. All the samples were formalin-fixed, paraffin-embedded, and immunostained with PDZRN4 antibody (ThermoFisher Scientific, Waltham, MA, USA). Quantification was conducted by counting the positive cells per field. Data is shown as mean ± standard deviation (SD).
Western blot
Cells were lysed with sodium dodecyl sulfate (SDS) lysis buffer (Beyotime, China) and denaturalized in a water bath at 100 °C. The prepared protein was resolved via SDS-PAGE, transferred to a polyvinylidene difluoride membrane from Millipore, and incubated with PDZRN4 primary antibody and HRP-Goat Anti-Rabbit IgG (H + L) (MultiSciences, Hangzhou, China). Protein levels were detected with a peroxide LumiGLO reagent (Cell Signaling Technology, 7003). Quantification of the relative protein levels was conducted according to the ratio of gray-value to the corresponding internal references. A representative graph was constructed and protein was quantified based on five pairs of clinical samples or three independent experiments and shown as mean ± SD.
Reverse transcription followed by real-time quantitative PCR (RT-qPCR)
RNA was extracted in accordance with the instructions for TRIzol reagent use (GENEray, Shanghai, China). The total RNA was subjected to reverse transcription using a kit from GENEray, Shanghai, China. Finally, cDNA was used for qPCR analysis. The RNA levels were normalized to internal references, and shown as mean ± SD (for primers, Table 1).
Table 1
| Primers | Sequences |
|---|---|
| PDZRN4-forward | TTTGCCCTGGAGCGCTTCGCAGA |
| PDZRN4-reverse | CGCCTCCAGCTCGTGCAGCCTGA |
| miR-221-3p-forward | CAAGGAATCATGTATGCTGTAG |
| miR-221-3p-reverse | AGGATGACATTACACCTTATCTC |
| U6-forward | GCTTCGGCAGCACATATACTAAAAT |
| U6-reverse | CGCTTCACGAATTTGCGTGTCAT |
Fluorescence-activated cell sorting (FACS, flow cytometry)
The rate of GFP-positive cells was determined using Accµri C6 (BD, San Jose, CA, USA). Cells were digested into single cells and determined by the GFP channel of Accµri C6. GFP-positive cells and the total cell number were counted automatically. The percentage of positive cells was calculated as positive cells number/total cell number ×100%.
Cell proliferation assay using cell-counting kit 8 (CCK8)
Two thousand cells were suspended in 100-µL culture medium and seeded per well in a 96-well plate; 8 plates were seeded. Each plate was collected daily after seeding, 10 µL CCK8 solution (MultiSciences, Hangzhou, China) was added, and measurements were performed using a microplate reader at 450 nm; the first measurement was done a few hours after seeding. The relative proliferation rate is shown as mean ± SD of reads at 450 nm.
Plate clone formation assay
One hundred cells were suspended in 2 mL culture medium in a 6-well plate. The medium was replenished every 2 days. The cell culture was observed daily until visible clones had formed. The culture medium was carefully removed and clones were stained with crystal violet solution. Data is shown as mean ± SD of numbers of clones per field.
Migration and invasion assay
Cells were seeded to the upper chamber of Millicell hanging cell culture inserts (Merck Millipore, Darmstadt, Germany). Culture medium without FBS and complete culture medium, were added, respectively, to the upper and lower chambers of the culture wells. Cells that had migrated to the lower surface of the filter were fixed with 70% methanol and then stained with 0.5% crystal violet solution.
For the invasion assay, hanging cell culture inserts were pre-coated with diluted Matrigel an hour before cell seeding. The cells that migrated or invaded the lower surface of the wells were counted. Data are shown as mean ± SD. Student’s t-test was performed when comparing data between groups.
Wound-healing assay
Cells were seeded to six-well culture plates and cultured overnight. By the second day, the culture medium was carefully removed and medium pipette tips were used to scratch the monolayer cell culture. Floating cells were carefully removed and fresh medium was added to the cell culture.
Graphs were constructed at the time of the scratch (0 hour) and 12 hours after treatment.
The percentage of scratch remaining after wounding was calculated as: (measurement at 12 hours/measurement at 0 hour) ×100%. The percentage of wound closure was calculated as: 100% − percentage of wound remaining. The percentage of wound closure is shown as mean ± SD.
Statistical analysis
The data in this study are shown as mean ± SD. The comparison of means between two groups was conducted using Student’s t-test. P<0.05 was considered significant; *P<0.05, **P<0.01 and ***P<0.001.
Results
PDZRN4 is downregulated in colon cancer at both the mRNA and protein level
In the initial phase of this investigation, mRNA array data obtained from NCBI was analyzed (GEO accession: GSE75970 and GSE74604). We compared gene expression levels in both the array and screened differentially expressed gene, genes having a 2-fold difference in quantity between colon cancer tissues and normal tissues (Figures S1,S2). After searching studies and reports concerning differentially expressed genes, we found PDZRN4, downregulated in both arrays, had never been studied in colon cancer, and was not well illustrated in almost any disease.
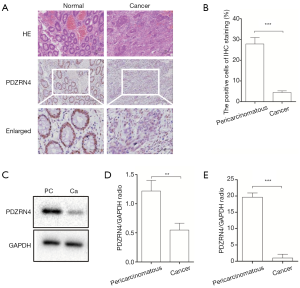
The levels of PDZRN4 protein in 5 pairs of colon cancer tissues and matched normal tissues were compared using IHC and western blot (Figure 1A,B,C,D). Both analyses showed lower amounts of PDZRN4 protein in the colon cancer tissues than in the normal tissues. In addition, RT-qPCR of 20 pairs of samples showed less PDZRN4 mRNA in the cancer samples (Figure 1E). These results suggest that PDZRN4 is suppressed at both the mRNA and translated levels in colon cancer.
Ectopic expression of PDZRN4 attenuated HCT116 cell’s tumorigenesis
To determine if PDZRN4 may be a regulator in colon cancer development, the gene was overexpressed in HCT116 cells (Figure 2A). The FACS, western blot, and RT-qPCR results confirmed the successful overexpression of PDZRN4 (Figure 2B,C,D, respectively).
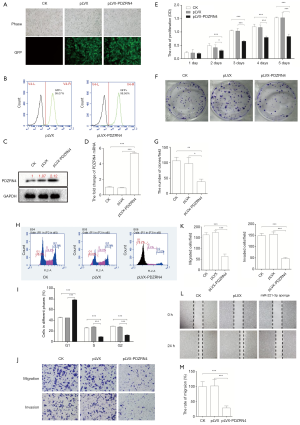
To investigate how PDZRN4 regulates cancer cell activities, the cells overexpressing PDZRN4 were then applied for functional assays. The cells overexpressing PDZRN4 proliferated less compared with the control cells, as measured by the cell proliferation (Figure 2E) and plate clone formation (Figure 2F,G) assays. A study of the cell cycle of the transformed cells showed that fewer of those overexpressing PDZRN4 reached S phase, compared with the control. This suggests that there was less DNA replication in the overexpressing cells (Figure 2H,I).
Tumor cells often have an enhanced ability to disseminate. In the present study, cells with overexpressed PDZRN4 were much less able to disseminate via migration and invasion (Figure 2J,K), as also indicated by the wound healing assay (Figure 2L,M). Altogether, these data suggest that PDZRN4 is suppressed in colon cancer, and this suppression promotes cancer cell proliferation and dissemination. Thus, PDZRN4 may function as a tumor suppressor.
With this supposition, the regulation of PDZRN4 expression in colon cancer was investigated.
PDZRN4 mRNA is a direct target of miR-221-3p
Since PDZRN4 expression was dysregulated at both the mRNA and protein levels, we investigated whether it may be regulated by miRNAs. Thus, data from the noncoding RNA expression profile (GEO accession: GSE101501) was explored for implications in colon cancer. Using the databases RNAhybrid, TargetScanHuman, miRWalk, and miR, the following miRNAs listed as upregulated in colon cancer tissues in the databases were selected and investigated for binding potential with the PDZRN4 3'-UTR: miR-153-3p, miR-5195-3p, miR-371a-3p, miR-133a-5p, miR-221-3p, miR-31-5p, miR-373-3p, miR-615-5p, miR-582-3p, and miR-301b-3p. To determine the association of these miRNAs with PDZRN4, PDZRN4 was measured after knocking out each miRNA respectively. It was found that, among these miRNAs, transfection with the miR-221-3p inhibitor was associated with dramatically higher levels of both protein PDZRN4 and mRNA PDZRN4 (Figure 3A,B).
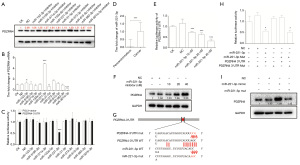
To verify binding further, luciferase report assays were conducted. The results showed inhibition of luciferase activity, indicating that miR-221-3p directly binds to the 3'-UTR of PDZRN4 mRNA (Figure 3C).
Further investigation revealed that, compared with normal tissues, miR-221-3p is overexpressed in colon cancer tissues (Figure 3D). At this point in the investigation, to confirm the presence of a regulation network, the miR-221-3p and PDZRN4 3'-UTR vector was transfected into 293T cells. The luciferase report assay showed that luciferase activity was dose-dependent with miR-221-3p (Figure 3E), and this was true also for PDZRN4 protein based on the western blot (Figure 3F). These results indicated the direct inhibition of PDZRN4 expression by miR-221-3p.
To specify the binding sequence, we mutated several bases in both miR-221-3p and the PDZRN4 3'-UTR (Figure 3G). According to the dual luciferase assay, mutation in either miRNA or the PDZRN4 3'-UTR attenuated regulation (Figure 3H), and western blot was consistent with this (Figure 3I). These data indicated that the mutant regions of miR-221-3p and the PDZRN4 3'-UTR are the binding sites of this miR-3' UTR regulation system (Figure 3G).
Knockdown of miR-221-3p restrained tumorigenesis of HCT116 cells
To support that miR-221-3p-regulated PDZRN4 expression interfered with the activity of colon cancer cells, miR-221-3p was knocked down by transducing HCT116 cells with a miRNA sponge. Observation and FACS detection of red fluorescent protein revealed that miR-221-3p was successfully dysregulated in HCT116 cells (Figure 4A,B). In addition, consistent with the results of miRNA overexpression, the levels of PDZRN4 protein were comparable with that observed prior to knockdown of miR-221-3p (Figure 4C).
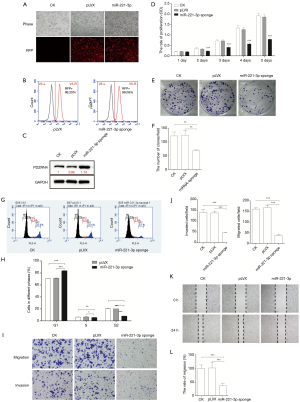
We then conducted more assays of phenotype. Both the cell proliferation (Figure 4D) and plate clone formation (Figure 4E,F) assays indicated that suppression of miR-221-3p expression lowered the proliferation of HCT116 cells. In addition, observations of the cell cycle showed inhibition of cell cycle progression (Figure 4G,H). Together, these data indicate that miR-221-3p functions in regulating HCT116 cell proliferation.
Moreover, the transwell migration, invasion, and wound-healing assays showed intense inhibition of cancer cell dissemination after treatment with the miR-221-3p sponge (Figure 4I,J,K,L). Together, these data indicate that miR-221-3p modulated the dissemination of colon cancer HCT116 cells.
Discussion
Gene dysregulation is a feature of cancer development. Various dysregulated genes may function as either tumor suppressors or oncogenes. Thus, we searched and analyzed gene expression profiles, and chose an inhibited gene in colon cancer for further investigation, i.e., PDZRN4. The PDZRN4 mRNA and PDZRN4 protein levels in colon cancer tissues were much lower than that of the adjacent normal tissues. Overexpressed PDZRN4 in the colon cancer cell line arrested cell cycle, inhibited cell proliferation, and attenuated migration and invasion. This suggests that PDZRN4 protein in colon cancer tissues is negatively associated with cancer cell dissemination, and PDZRN4 may function as a tumor suppressor in colon cancer development.
Furthermore, we found that PDZRN4 translation was inhibited by an overexpressed onco-miRNA, miR-221-3p. The luciferase report assay showed a direct association between miR-221-3p and the 3'-UTR of PDZRN4 mRNA. In addition, the cell proliferation and plate clone formation assays showed that as the miR-221-3p level subsided, so did the proliferation and dissemination of the colon cancer cell. This indicates that miR-221-3p-PDZRN4 has an important role in regulating colon cancer development, in which miR-221-3p inhibits the translation of PDZRN4 and thereby promotes the proliferation and dissemination of colon cancer cells.
In this study, the low number of clinical samples and time allowed limited sufficient inspection PDZRN4 expression in colon cancer tissues. Whether PDZRN4 protein is a viable biomarker of colon cancer requires further investigation.
Conclusions
This study investigated the expression of PDZRN4 in colon cancer tissues, its potential function in the development of colon cancer, and its inhibition by its upstream regulator, miR-221-3p. Three main conclusions are highlighted. First, according to the gene expression profile and detection in clinical colon cancer samples, PDZRN4 mRNA and PDZRN4 protein are downregulated in colon cancer. Second, overexpression of PDZRN4 in the colon cancer cell line and the phenotype assays indicated that PDZRN4 functions to attenuate cell proliferation, migration, and invasion. Thirdly, miR-211-3p inhibited PDZRN4 translation by binding to its mRNA 3'-UTR. The knockdown of miR-211-3p dramatically reduced the proliferation, migration, and invasion of colon cancer HCT116 cells.
These discoveries suggest that PDZRN4 may be a potential target for defeating colon cancer, but more investigations are necessary.
Table S1
| Patient No. | Gender | Age, years | Histological type | LNM | Tumor size, cm3 |
|---|---|---|---|---|---|
| PT 1 | F | 64 | Mucinous adenocarcinoma | POS | 3.5×4.5×1.5 |
| PT 2 | M | 75 | Canalicular adenoma | POS | 8.0×5.5×2.5 |
| PT 3 | F | 83 | Signet-ring cell carcinoma | POS | 7.0×4.5×2.0 |
| PT 4 | M | 79 | Mucinous adenocarcinoma | POS | 5.0×4.0×2.3 |
| PT 5 | M | 63 | Canalicular adenoma | POS | 3.5×2.0×1.0 |
| PT 6 | F | 37 | Canalicular adenoma | POS | 5.0×4.5×2.0 |
| PT 7 | F | 85 | Canalicular adenoma | NEG | 4.0×4.0×1.0 |
| PT 8 | M | 51 | Signet-ring cell carcinoma | POS | 6.0×2.5×2.0 |
| PT 9 | F | 84 | Canalicular adenoma | NEG | 10.0×5.0×4.5 |
| PT 10 | M | 73 | Canalicular adenoma | NEG | 5.5×3.5×2.0 |
| PT 11 | M | 74 | Canalicular adenoma | POS | 4.0×3.3×1.5 |
| PT 12 | F | 55 | Canalicular adenoma | NEG | 1.1×0.9×0.5 |
| PT 13 | F | 58 | Canalicular adenoma | POS | 6.5×4.0×3.0 |
| PT 14 | F | 74 | Mucinous adenocarcinoma | POS | 3.5×2.0×1.5 |
| PT 15 | M | 64 | Polypoid adenocarcinoma | POS | 3.0×2.2×1.6 |
| PT 16 | M | 60 | Canalicular adenoma | POS | 4.3×3.2×1.5 |
| PT 17 | F | 70 | Canalicular adenoma | POS | 8.0×5.0×2.5 |
| PT 18 | F | 58 | Canalicular adenoma | POS | 6.0×3.5×2.0 |
| PT 19 | M | 71 | Canalicular adenoma | POS | 3.2×3.0×0.8 |
| PT 20 | M | 78 | Canalicular adenoma | POS | 6.0×4.0×4.0 |
F, female; LNM, lymph node metastasis; M, male; NEG, negative; POS, positive; PT, patient.
Acknowledgments
Funding: This research was supported by
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.07.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Institutional Ethics Committee of Second Affiliated Hospital of Soochow University (Suzhou, 215004, Jiangsu Province, China) reviewed and authorized the clinical section of this research. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brouwer NPM, Bos ACRK, Lemmens VEPP, et al. An overview of 25 years of incidence, treatment and outcome of colorectal cancer patients. Int J Cancer 2018;143:2758-66. [Crossref] [PubMed]
- Zhang Y, Shi J, Huang H, et al. Burden of colorectal cancer in China. Zhonghua Liu Xing Bing Xue Za Zhi 2015;36:709-14. [PubMed]
- Poelemeijer YQM, Lijftogt N, Detering R, et al. Obesity as a determinant of perioperative and postoperative outcome in patients following colorectal cancer surgery: A population-based study (2009-2016). Eur J Surg Oncol 2018;44:1849-57. [Crossref] [PubMed]
- Pang Y, Kartsonaki C, Guo Y, et al. Diabetes, plasma glucose and incidence of colorectal cancer in Chinese adults: a prospective study of 0.5 million people. J Epidemiol Community Health 2018;72:919-25. [Crossref] [PubMed]
- Stein L, Rothschild J, Luce J, et al. Copy number and gene expression alterations in radiation-induced papillary thyroid carcinoma from chernobyl pediatric patients. Thyroid 2010;20:475-87. [Crossref] [PubMed]
- Hua Y, Ma X, Liu X, et al. Abnormal expression of mRNA, microRNA alteration and aberrant DNA methylation patterns in rectal adenocarcinoma. PLoS One 2017;12:e017446. [Crossref] [PubMed]
- Hu T, Yang H, Han ZG. PDZRN4 acts as a suppressor of cell proliferation in human liver cancer cell lines. Cell Biochem Funct 2015;33:443-9. [Crossref] [PubMed]
- Andres SF, Williams KN, Plesset JB, et al. IMP1 3' UTR shortening enhances metastatic burden in colorectal cancer. Carcinogenesis 2019;40:569-79. [Crossref] [PubMed]
- Wang T, Hou J, Jian S, et al. miR-29b negatively regulates MMP2 to impact gastric cancer development by suppress gastric cancer cell migration and tumor growth. J Cancer 2018;9:3776-86. [Crossref] [PubMed]
- Yu L, Xiang L, Feng J, et al. miRNA-21 and miRNA-223 expression signature as a predictor for lymph node metastasis, distant metastasis and survival in kidney renal clear cell carcinoma. J Cancer 2018;9:3651-9. [Crossref] [PubMed]
- Xie X, Huang Y, Chen L, et al. miR-221 regulates proliferation and apoptosis of ovarian cancer cells by targeting BMF. Oncol Lett 2018;16:6697-704. [PubMed]
- Yang Y, Li H, Ma Y, et al. MiR-221-3p is down-regulated in preeclampsia and affects trophoblast growth, invasion and migration partly via targeting thrombospondin 2. Biomed Pharmacother 2019;109:127-34. [Crossref] [PubMed]
- Zheng X, Dai J, Zhang H, et al. MicroRNA-221 promotes cell proliferation, migration, and differentiation by regulation of ZFPM2 in osteoblasts. Braz J Med Biol Res 2018;51:e7574. [Crossref] [PubMed]
- Cai K, Shen F, Cui JH, et al. Expression of miR-221 in colon cancer correlates with prognosis. Int J Clin Exp Med 2015;8:2794-8. [PubMed]
- Tao K, Yang J, Guo Z, et al. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res 2014;6:391-401. [PubMed]
- Liu S, Sun X, Wang M, et al. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFκB and STAT3 in colorectal cancer cells. Gastroenterology 2014;147:847-59.e11. [Crossref] [PubMed]
- Tsunoda T, Takashima Y, Yoshida Y, et al. Oncogenic KRAS regulates miR-200c and miR-221/222 in a 3D-specific manner in colorectal cancer cells. Anticancer Res 2011;31:2453-9. [PubMed]
- Moshiri F, Callegari E, D'Abundo L, et al. Inhibiting the oncogenic mir-221 by microRNA sponge: toward microRNA-based therapeutics for hepatocellular carcinoma. Gastroenterol Hepatol Bed Bench 2014;7:43-54. [PubMed]