
The expression profile of RNA sensors in colorectal cancer and its correlation with cancer stages
Introduction
Colorectal cancer (CRC), one of the most common gastrointestinal malignancy with high morbidity and mortality, poses a serious threat to human health, especially for people in developing or underdeveloped countries (1). Most of patients have already in the advanced stages when they are diagnosed as CRC, and therefore resection and postoperative chemotherapy are often required. As a result, the living quality of these patients is seriously affected. A better understanding on the underlying mechanisms and the identification of molecular candidates more suitable for CRC diagnosis, prognosis, and treatment are needed. The pathogenesis of CRC is complicated, and the interactions among gut microbiota, immune system, diets, and environment may all be involved (2). Among them, inflammation is a well-known risk factor for CRC tumorigenesis, and is often initiated by pattern recognition receptors (PRRs)-induced cascade response in immune cells (3).
Nucleic acids (DNA and RNA) represent important members of pathogen-associated molecular patterns (PAMPs) derived from pathogens, and the innate immune system employ a range of nucleic acid sensors (including DNA sensors and RNA sensors, subsets of PRRs) to detect various DNA and/or RNA components from these pathogens, thus restricting their replication (4,5). Previous studies have shown that nucleic acid sensors also respond to self-derived nucleic acids, known as damage-associated molecular patterns (DAMPs). The activation of DNA sensors is involved in inflammatory responses and tumorigenesis, and these sensors have prognosis and therapeutic values in some cancers. Furthermore, many DNA sensors were dysregulated in CRC and might be associated with cancer stages (4).
Similar to DNA sensors, RNA sensors are also important to restrict viral infection especially for viruses with rapid replication capability and high mutation rates (6). The members of RNA sensors mainly include endosomal receptors such as Toll-like receptor 3/7/8 (TLR3/7/8), cytosolic receptors such as RIG-I like receptor (RLRs) (including Retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene (MDA5), and DExH-box helicase 58 (DHX58, also named as LGP2), and helicases such as DDX1 and DDX21 (5). Upon activation by corresponding RNA components, RNA sensors signal via different adaptors (TRIF for TLR3, MyD88 for TLR7/8, and MAVS for RLRs) and eventually lead to the production of type I interferons (type I IFNs, IFN-α/β), other inflammatory cytokines and chemokines, which are important for the activation of innate and adaptive immune responses (5). Similar to DNA sensors, aberrant activation of RNA sensors are involved in the development of chronic inflammatory diseases such as autoimmunity diseases and tumorigenesis (3,6,7). In addition, deregulation of RNA sensors is involved in the development of CRC or inflammatory bowel diseases (IBDs).
The RNA-sensing TLRs are the most studied RNA sensors in intestinal diseases. They are one of the major drivers for NF-κB activation, yet their roles in CRC are still unclear (8-10). RIG-I and MDA5 are the two main members of RLRs with similar structures (11), but they respond to dsRNA with different lengths (12). A previous study showed that Rig-I-/- mice were more susceptible to azoxymethane (AOM)/dextran sodium sulfate (DSS)-induced CRC, which may be partially associated with a disturbance of gut microbiota (13,14). MDA5 was reported to protect mice from the development of various cancers (4,14). Different from RIG-I and MDA5, LGP2, as another member of RLRs, lacks the N-term cascade domain and mainly acts as a positive or negative regulator for MDA5 and RIG-I (15). A study showed that LGP2 was important for the survival of CRC cell line receiving ionizing radiation (16). While the other two members of RNA sensors, DDX1 and DDX21, were reported to interact with DHX36 and mediate the production of type I IFNs via TRIF (17), the roles of DDX1 and DDX21 in CRC development have yet to be studied.
These previous studies suggested that RNA sensors might be involved in the development of CRC. However, the expression profile of RNA sensors in human CRC and their clinical significance as well as the underlying mechanisms are still largely unknown. Here, we systematically investigate the expression profile of RNA sensors in human CRC tissues and mouse colon tissues at early stage of AOM/DSS-induced CRC. Furthermore, we take advantage of Oncomine bioinformatics platform to analyze the expression patterns of RNA sensors in a large scale and explore the possible functional mechanisms. The present study may have a significant value for the diagnosis, prognosis, and therapy of CRC.
Methods
Clinical sample collection
In this study, 34 CRC tissues and matched peri-carcinomatous tissues (distance (3 cm from the tumor sites) were obtained between July 2015 and April 2017 from Department of Gastrointestinal Surgery of the First Affiliated Hospital of Gannan Medical University (Ganzhou, Jiangxi, China). Written informed consents were obtained from all participants. The present study was approved by the Ethics Committee of Gannan Medical University. Patients who were newly diagnosed with CRC based on the clinicopathological criteria were included. Patients who had received preoperative chemo-, radio- or immunotherapy were excluded. After the excision of CRC, the tissues were collected and preserved in liquid nitrogen. All information of the patients includes gender, age, tumor size, the stages and grades of the tumor were recorded (Table S1).
AOM/DSS treatment of mice
AOM/DSS-induced colitis-associated CRC model is often used to analyze the relationship between innate immune receptors and CRC (18). AOM is a pro-carcinogen to induce gene mutation, and DSS is an inflammatory insult to induce inflammation. Briefly, 6–8 weeks old C57BL/6 mice (n=5, Model Animal Research Center of Nanjing University, Nanjing, China) were intraperitoneally injected with AOM (Sigma, USA, 10 µg/g body weight) followed by regular drinking water for 5 days, then the mice were fed up with drinking water containing 3% DSS (MP Biologicals, USA) for 6 days. After that, the mice gained full access to regular drinking water for 3 days (14 days). In contrast, the control group mice (n=7) were intraperitoneally injected with PBS and fed up with regular drinking water throughout the experiment. Mice were sacrificed when they lost more than 25% of body weight for humane endpoints. In this study, we scarified the mice using CO2 at the day 14 after AOM injection, a time point that is frequently used to study the pre-cancerous changes and analyze the CRC pathogenesis (19-21). In order to test the TLR expression levels after the development of colitis, C57BL/6 mice were fed up with drinking water containing 3% DSS for 6 days, and then the mice got full access to regular drinking water for 3 days (day 9). After that, mice were sacrificed for colon collection.
qRT-PCR
The total RNA was isolated from human and mouse colon tissues by Trizol reagents according to manufacturer’s instructions (Invitrogen, USA). After that, the cDNA was synthesized by using M-MLV First Strand Kit (Invitrogen, USA). The real-time quantitative PCR was performed by using SYBER Green Master Mix (Invitrogen, USA) in Bio-Rad Real-Time PCR System. The primer sequences of RNA sensors and β-actin were listed (Tables S2,S3).
Data collection from Oncomine® Bioinformatics platform
Oncomine (http://www.oncomine.org) is a unique resource platform that delivers unified cancer microarray data from various databases (22). In the present study, the mRNA levels of RNA sensors in human CRC were analyzed in TCGA database since most CRC samples were collected from this database. The data we collected for further analysis met the following requirements: Analysis Type: Cancer vs. Normal Analysis; Cancer Type: Colorectal Cancer; Data Type: mRNA; Sample Type: Clinical Specimen; Genes: RNA sensors; Dataset name: TCGA Colorectal, Threshold fold change ≥1.5, P value ≤10−4, Gene Rank in the top 10). After the primary data were collected, they were organized by sample type (Cancer vs. Normal), tumor stages, and patients ID to meet different analysis requirements.
Statistical analysis
All the statistical analysis was performed in Prism 7.0 software, and data were presented as mean ± SEM. The relative expression of mRNA in our clinical CRC samples and mouse colon tissues at early stage of CRC formation was obtained by 2−ΔΔCT. The expression difference between clinical CRC tissues and adjacent tissues were analyzed by pair Student’s t-test. Unpaired t test was used to analyze the change between CRC and health control in TCGA database and AOM/DSS-induced CRC or the control group. In addition, the changes of RNA sensors in different cancer stages were determined by analysis of variance (ANOVA) with Dunnett’s post-hoc analysis. Finally, the expression correlation between RNA sensors and the main adaptors were analyzed by multivariate linear regression analysis (Liner, regression). P<0.05 was considered statistically significant. *P<0.05, **P<0.01, and ***P<0.001.
Results
RNA-sensing TLRs were dysregulated in CRC tissues and associated with cancer stages
RNA-sensing TLRs (TLR3/7/8) are the most studied RNA sensors in intestinal diseases although their roles in CRC are still controversial. An overall understanding on their expression profiles in the CRC tissues is beneficial for clinical application. We initially analyzed the expression profiles of TLR3/7/8 in clinical CRC samples, colon tissues from AOM/DSS treated mice, and TCGA database. We found that the expression of all these genes significantly decreased in the samples of three different groups in comparison with controls (Figure 1).
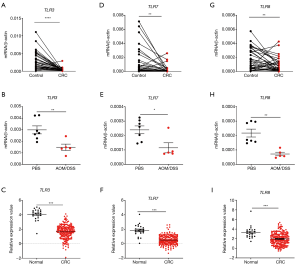
In order to investigate whether all TLRs were down-regulated in CRC development, the gene expression levels of other TLRs were determined in mouse colon tissues after AOM/DSS treatment. It was shown that at least TLR2 and TLR4 expression levels were not significantly different between control and AOM/DSS-treated groups (Figure S1A,B). It indicates that the down-regulation of TLRs in CRC may be specific for RNA-sensing TLRs.
Furthermore, in order to determine whether RNA-sensing TLRs are downregulated in CRC development or in any inflammatory conditions, we measured the gene expression levels of TLR3/7/8 in mouse colon tissues after DSS treatment only. It was shown that TLR3/7/8 were not significantly down-regulated after DSS only treatment (Figure S1C,D,E). It indicates that the down-regulation of RNA-sensing TLRs is specific for CRC but not for other inflammatory conditions.
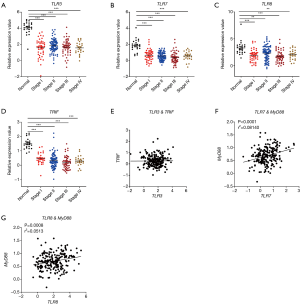
Next, we analyzed the correlation between the expression level of the TLRs and cancer stages in TCGA database. Consistently, we found that the expression levels of both TLR3/7/8 (Figure 2A,B,C) and the adaptors TRIF (Figure 2D) in different stages of CRC were significantly different from healthy control group. We also analyzed the possible signaling pathways of RNA-sensing TLRs in CRC development. We found that, although TLR3 showed no correlation with TRIF, there was a positive correlation between TLR7/8 and MyD88 (Figure 2E,F,G). These results showed the correlation of the downregulation of genes encoding RNA-sensing TLRs and their adaptor molecules with the development of CRC.
RLRs were dysregulated in CRC and associated with cancer stages
RLRs include MDA5, RIG-I, and LGP2. RIG-I and MDA5 mainly signal via the adaptor MAVS and induce the production of type I IFNs. In our study, we found that RIG-I was down-regulated in human and mouse colon tissues and in TCGA database (Figure 3A,B,C). Similarly, MDA5 was down-regulated in human and mouse colon tissues (Figure 3D,E). However, no significant change of MDA5 was observed in TCGA database (Figure 3F). In addition, MAVS was up-regulated in CRC in TCGA database (Figure 3G). Furthermore, it was found that the expression of RIG-I in CRC was associated with cancer stages (Figure 4A), whereas the expression of MDA5 and MAVS showed no correlation with cancer stages (Figure 4B,C).
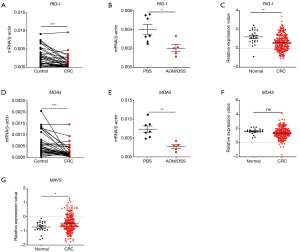
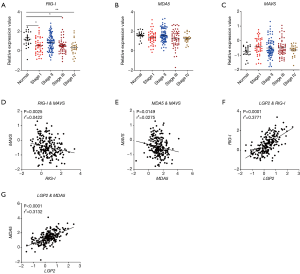
Next, we analyzed the expression correlation between RIG-I/MDA5 and the adaptor MAVS. Surprisingly, both the expression of RIG-I/MDA5 in CRC was negatively associated with MAVS (Figure 4D,E). Furthermore, we also determined the expression levels of type I IFNs (IFN-α/β) in CRC tissues and could not find the significant down-regulation of both IFN-α and IFN-β in CRC tissues (Figure S2A,B). These results indicate that the role of RLRs in CRC may be independent of classical RLR-MAVS-Type I IFNs signaling pathway, which is consistent with previous studies (14). On the other hand, as a member of RLRs, LGP2 lacks the N-term cascade domain and mainly acts as a positive regulator for MDA5 and a negative for RIG-I. Here we found that LGP2 was positively correlated with both RIG-I and MDA5 in CRC (Figure 4F,G), although underlying mechanism is unknown. Taken together, our study showed that RLRs were down-regulated in CRC tissues and might be potentially involved in CRC development.
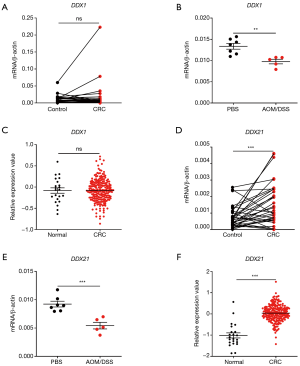
The RNA-sensing helicases DDX1 and DDX21 were deregulated in CRC and associated with cancer stages
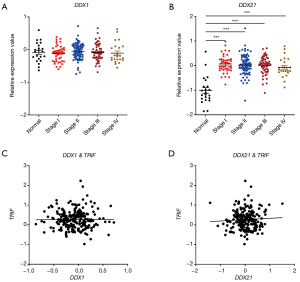
Next, we studied the expression patterns of the RNA sensing-helicases (DDX1 and DDX21) in CRC. We found that, although DDX1 expression was down-regulated in mouse colon tissues after AOM/DSS, it was not differentially expressed in clinical human CRC tissues and in TCGA database (Figure 5A,B,C). In contrast to DDX1, the expression of DDX21 significantly increased in human CRC tissues and TCGA database, although it was downregulated in mouse colon tissues after AOM/DSS treatment in comparison with controls (Figure 5D,E,F). In addition, DDX21 expression was associated with cancer stages, whereas DDX1 expression was not (Figure 6A,B). It is possible that the expression profile of DDX21 in CRC development is a dynamic process. DDX1 and DDX21 can interact with DHX36 and signal via TRIF to induce the production of type I IFNs (17). However, in our present study, we found that the expression of DDX1/DDX21 in CRC showed no correlation with TRIF (Figure 6C,D).
Discussion
Aberrant activation of inflammatory response represents a hallmark of tumorigenesis and the understanding on its underlying molecular mechanisms is of benefit for cancer diagnosis, prognosis, and therapy (23). Recent accumulating evidences showed that nucleic acid sensors (DNA sensors and RNA sensors) are not only the indispensable PRRs for pathogen detection and elimination but also the main initiators of inflammation (6). Moreover, recent studies extended the function of nucleic acid sensors to tumorigenesis in addition to pathogen restriction and inflammation development (4,24). However, the expression profile of these nucleic acid sensors in CRC has not been well characterized.
Interaction between TLRs and tumor cells are complicated and TLRs appear to have tumor-promoting effects in many cancers (25). However, RNA sensing TLRs seems to have conflicting roles in the development of cancers. On one hand, TLR3 can promote the invasiveness of metastatic CRC cell lines by enhancing the production of pro-inflammatory cytokines (26). On the other hand, TLR3 was shown to directly induce apoptosis in human breast cancer cells and breast cancer patients with loss of function allele in TLR3 exhibit reduced survival rates after chemotherapy (27,28). Furthermore, the specific TLR3 ligand induced activation of NK cells and CD8+ T cells and established potent anti-tumor immunity in mouse tumor models (29,30). Here, we found that the expression of TLR3 was reduced in clinical CRC samples, TCGA database, and mouse colon tissues, and associated with cancer stages, indicating the potential anti-tumor effect of TLR3.
A study showed that the expression of TLR7 and TLR8 were higher in tumor tissues and might be associated with a poorer survival (31). However, more recent studies suggest that activation of TLR7 and TLR8 by various ligands induced tumor regress in various tumor models including CRC (32-36). Our present study showed lower expression of TLR7 and TLR8 in CRC tissues in comparison with controls, indicating potential anti-tumor effects.
RLRs, another subfamily of RNA sensors, include RIG-I, MDA5, and LGP2. Multiple previous studies have showed that RLRs were indispensable for RNA virus restriction, and the lack of MDA5, RIG-I and MAVS may increase virus proliferation in intestinal epithelial cells, such as rotavirus (37,38). Consistently, previous animal studies have also shown that RIG-I, MDA5, and MAVS inhibited the development of DSS-induced colitis (4). A recent study also showed that RIG-I suppressed the development of AOM/DSS-induced CRC (13). Encouragingly, activation of RIG-I may promote the integrity of intestinal epithelium suffering irradiation treatment and reduce graft-versus-host disease (39). These indicated that RLRs may be involved in the development of CRC, and the understanding on their expression profile in CRC may not only be of prognosis value but also be of therapy value. Our present study found that RIG-I and MDA5 were significantly down-regulated in CRC. Moreover, the expression of RIG-I in CRC was associated with cancer stages and LGP2 expression positively correlated with the expression of RIG-I and MDA5. These results are in line with previous literature (14). However, further functional investigations of RLRs in CRC development and therapy are still needed.
The helicase family is a large protein family that is expressed in almost all tissues and cells. They mainly hydrolyze RNA double chains in an energy-dependent manner throughout the whole process of life (40). Previously, studies on the function of helicase family members are mainly focused on viral defense, in addition to organ development processes. In recent years, these family members were shown to be functionally implicated in the development of cancers, including CRC (41). Recent studies showed that DDX1 inhibited ovarian cancer progression through promoting microRNA maturation, yet promoted CRC through transcription activation of LGR5 (42,43). DDX21 was shown to promote the development of breast cancer and gastric cancer and was elevated in CRC (44-46). Here, consistent with previous studies, we showed that the expression of DDX21 were up-regulated in CRC and associated with cancer stages. However, the possible signaling pathway of DDX1 and DDX21 in CRC development remains undefined.
Taken together, our present study facilitates the comprehensive understanding on the expression patterns, clinical significance, and possible roles of RNA sensors in CRC. More importantly, we have identified that, among all these RNA sensors, TLR3/7/8, RIG-I, MDA5, and DDX21, may be potentially important for diagnosis, prognosis, and therapy of CRC, and therefore deserve further investigation in the near future.
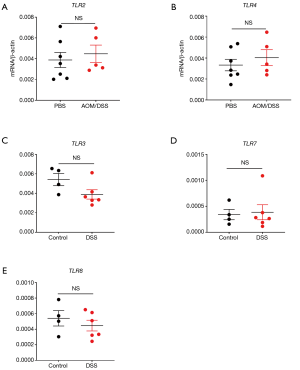
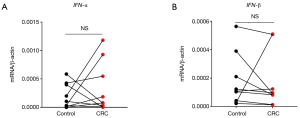
Table S1
| Parameters | Number (%) |
|---|---|
| Gender | |
| Male | 16 (47.1) |
| Female | 18 (52.9) |
| Age (years) | |
| <50 | 7 (20.6) |
| ≥50 | 27 (79.4) |
| Location | |
| Colon | 18 (52.9) |
| Rectal | 16 (47.1) |
| Clinical stages | |
| II | 16 (47.1) |
| III | 13 (38.2) |
| IV | 5 (14.7) |
| Grades | |
| 1 | 9 (26.5) |
| 2 | 20 (58.8) |
| 3 | 5 (14.7) |
| Pathologic type | |
| Adenocarcinoma | 28 (82.4) |
| Mucinous adenocarcinoma | 6 (17.6) |
CRC, colorectal cancer.
Table S2
| Genes | Sequences (5'-3') |
|---|---|
| DHX36 | (F): GGG TCA TGG AGG TAA CCG AG |
| (R): CTC TCC GCT TCC TTG TTC TTC | |
| TLR3 | (F): TTG CCT TGT ATC TTT TGG GG |
| (R): TCA ACA CTG TTA TGT TTG TGG GT | |
| TLR7 | (F): TCC TTG GGG CTA GAT GGT TTC |
| (R): TCC ACG ATC ACA TGG TTC TTT G | |
| TLR8 | (F): ATG TTC CTT CAG TCG TCA ATG C |
| (R): TTG CTG CAC TCT GCA ATA ACT | |
| MDA5 | (F): TAG AAT GGG TAT TCC ACA GAC G |
| (R): GTG GCG ACG GTC CTC TGA A | |
| RIG-I | (F): CTG GAC CCT ACC TAC ATC CTG |
| (R): GGC ATC CAA AAA GCC ACG G | |
| DDX1 | (F): TCT CCG AGA TGG GTG TAA TGC |
| (R): ACC TCC TCC TAA GAT CAA TGG G | |
| DDX21 | (F): GAG GCC GTT TCC TCC AAA G |
| (R): GTA GAA GCA GTG TCG TCT TGA G | |
| β-actin | (F): CTC CTT AAT GTC ACG CAC GAT |
| (R): CAT GTA CGT TGC TAT CCA GGC |
Table S3
| Genes | Sequences (5'-3') |
|---|---|
| TLR3 | (F): GTG AGA TAC AAC GTA GCT GAC TG |
| (R): TCC TGC ATC CAA GAT AGC AAG T | |
| TLR7 | (F): ATG TGG ACA CGG AAG AGA CAA |
| (R): GGT AAG GGT AAG ATT GGT GGT G | |
| TLR8 | (F): GAA AAC ATG CCC CCT CAG TCA |
| (R): CGT CAC AAG GAT AGC TTC TGG AA | |
| MDA5 | (F): AGA TCA ACA CCT GTG GTA ACA CC |
| (R): CTC TAG GGC CTC CAC GAA CA | |
| RIG-I | (F): AAG AGC CAG AGT GTC AGA ATC T |
| (R): AGC TCC AGT TGG TAA TTT CTT GG | |
| DDX1 | (F): CTC CGA AAT GGG TGT TAT GCC |
| (R): GCC ATG AGT ACA TCC CCT CCT | |
| DDX21 | (F): GAG GCC GTT TCC TCC AAA G |
| (R): GTA GAA GCA GTG TCG TCT TGA G | |
| β-actin | (F): CAG CTT CTT TGC AGC TCC TT |
| (R): CAC GAT GGA GGG GAA TAC AG |
Acknowledgments
Funding: This work was supported by funds from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.07.45). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Human samples used in the present study were obtained from the patients with written informed consent. The present study was approved by the Ethics Committee of Gannan Medical University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66:683-91. [Crossref] [PubMed]
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383:1490-502. [Crossref] [PubMed]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010;140:805-20. [Crossref] [PubMed]
- He L, Chen Y, Wu Y, et al. Nucleic acid sensing pattern recognition receptors in the development of colorectal cancer and colitis. Cell Mol Life Sci 2017;74:2395-411. [Crossref] [PubMed]
- Gurtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol 2013;21:413-20. [Crossref] [PubMed]
- Smith S, Jefferies C. Role of DNA/RNA sensors and contribution to autoimmunity. Cytokine Growth Factor Rev 2014;25:745-57. [Crossref] [PubMed]
- Xu XX, Wan H, Nie L, et al. RIG-I: a multifunctional protein beyond a pattern recognition receptor. Protein Cell 2018;9:246-53. [Crossref] [PubMed]
- Bednarczyk M, Muc-Wierzgon M, Walkiewicz K, et al. Profile of gene expression of TLR-signaling pathways in colorectal cancer tissues. Int J Immunopathol Pharmacol 2017;30:322-6. [Crossref] [PubMed]
- Royse KE, Chen L, Berger DH, et al. Expression of pattern recognition receptor genes and mortality in patients with colorectal adenocarcinoma. Int J Mol Epidemiol Genet 2017;8:8-18. [PubMed]
- Li TT, Ogino S, Qian ZR. Toll-like receptor signaling in colorectal cancer: carcinogenesis to cancer therapy. World J Gastroenterol 2014;20:17699-708. [Crossref] [PubMed]
- Kato H, Fujita T. RIG-I-like receptors and autoimmune diseases. Curr Opin Immunol 2015;37:40-5. [Crossref] [PubMed]
- Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006;441:101-5. [Crossref] [PubMed]
- Zhu H, Xu WY, Hu Z, et al. RNA virus receptor Rig-I monitors gut microbiota and inhibits colitis-associated colorectal cancer. J Exp Clin Cancer Res 2017;36:2. [Crossref] [PubMed]
- Wu Y, Wu X, Wu L, et al. The anti-cancer functions of RIG-I like receptors, RIG-I and MDA5, and their applications in cancer therapy. Transl Res 2017;190:51-60. [Crossref] [PubMed]
- Hornung V. SnapShot: Nucleic acid immune sensors, part 2. Immunity 2014;41:1066-1066.e1. [Crossref] [PubMed]
- Widau RC, Parekh AD, Ranck MC, et al. RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation. Proc Natl Acad Sci USA 2014;111:E484-91. [Crossref] [PubMed]
- Zhang Z, Kim T, Bao M, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 2011;34:866-78. [Crossref] [PubMed]
- Snider AJ, Bialkowska AB, Ghaleb AM, et al. Murine Model for Colitis-Associated Cancer of the Colon. Methods Mol Biol 2016;1438:245-54. [Crossref] [PubMed]
- Zaki MH, Vogel P, Malireddi RK, et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell 2011;20:649-60. [Crossref] [PubMed]
- Karki R, Man SM, Malireddi RK, et al. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature 2016;540:583-7. [Crossref] [PubMed]
- Man SM, Zhu Q, Zhu L, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell 2015;162:45-58. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Qian BZ. Inflammation fires up cancer metastasis. Semin Cancer Biol 2017;47:170-6. [Crossref] [PubMed]
- He L, Xiao X, Yang X, et al. STING signaling in tumorigenesis and cancer therapy: A friend or foe? Cancer Lett 2017;402:203-12. [Crossref] [PubMed]
- Zeromski J, Kaczmarek M, Boruczkowski M, et al. Significance and Role of Pattern Recognition Receptors in Malignancy. Arch Immunol Ther Exp (Warsz) 2019;67:133-41. [Crossref] [PubMed]
- Bugge M, Bergstrom B, Eide OK, et al. Surface Toll-like receptor 3 expression in metastatic intestinal epithelial cells induces inflammatory cytokine production and promotes invasiveness. J Biol Chem 2017;292:15408-25. [Crossref] [PubMed]
- Salaun B, Coste I, Rissoan MC, et al. TLR3 can directly trigger apoptosis in human cancer cells. J Immunol 2006;176:4894-901. [Crossref] [PubMed]
- Vacchelli E, Enot DP, Pietrocola F, et al. Impact of Pattern Recognition Receptors on the Prognosis of Breast Cancer Patients Undergoing Adjuvant Chemotherapy. Cancer Res 2016;76:3122-6. [Crossref] [PubMed]
- Matsumoto M, Tatematsu M, Nishikawa F, et al. Defined TLR3-specific adjuvant that induces NK and CTL activation without significant cytokine production in vivo. Nat Commun 2015;6:6280. [Crossref] [PubMed]
- Damo M, Wilson DS, Simeoni E, et al. TLR-3 stimulation improves anti-tumor immunity elicited by dendritic cell exosome-based vaccines in a murine model of melanoma. Sci Rep 2015;5:17622. [Crossref] [PubMed]
- Grimm M, Kim M, Rosenwald A, et al. Toll-like receptor (TLR) 7 and TLR8 expression on CD133+ cells in colorectal cancer points to a specific role for inflammation-induced TLRs in tumourigenesis and tumour progression. Eur J Cancer 2010;46:2849-57. [Crossref] [PubMed]
- Dovedi SJ, Adlard AL, Ota Y, et al. Intravenous administration of the selective toll-like receptor 7 agonist DSR-29133 leads to anti-tumor efficacy in murine solid tumor models which can be potentiated by combination with fractionated radiotherapy. Oncotarget 2016;7:17035-46. [Crossref] [PubMed]
- Scholch S, Rauber C, Tietz A, et al. Radiotherapy combined with TLR7/8 activation induces strong immune responses against gastrointestinal tumors. Oncotarget 2015;6:4663-76. [Crossref] [PubMed]
- Scholch S, Rauber C, Weitz J, et al. TLR activation and ionizing radiation induce strong immune responses against multiple tumor entities. Oncoimmunology 2015;4:e1042201. [Crossref] [PubMed]
- Wang C, Zhou Q, Wang X, et al. The TLR7 agonist induces tumor regression both by promoting CD4(+)T cells proliferation and by reversing T regulatory cell-mediated suppression via dendritic cells. Oncotarget 2015;6:1779-89. [PubMed]
- Medler T, Patel JM, Alice A, et al. Activating the Nucleic Acid-Sensing Machinery for Anticancer Immunity. Int Rev Cell Mol Biol 2019;344:173-214. [Crossref] [PubMed]
- Chen N, Xia P, Li S, et al. RNA Sensors of the Innate Immune System and Their Detection of Pathogens. IUBMB Life 2017;69:297-304. [Crossref] [PubMed]
- Broquet AH, Hirata Y, McAllister CS, et al. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol 2011;186:1618-26. [Crossref] [PubMed]
- Fischer JC, Bscheider M, Eisenkolb G, et al. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci Transl Med 2017; [Crossref] [PubMed]
- Abdelhaleem M. Helicases: an overview. Methods Mol Biol 2010;587:1-12. [Crossref] [PubMed]
- Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol 2013;10:121-32. [Crossref] [PubMed]
- Han C, Liu Y, Wan G, et al. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep 2014;8:1447-60. [Crossref] [PubMed]
- Tanaka K, Ikeda N, Miyashita K, et al. DEAD box protein DDX1 promotes colorectal tumorigenesis through transcriptional activation of the LGR5 gene. Cancer Sci 2018;109:2479-89. [Crossref] [PubMed]
- Zhang Y, Baysac KC, Yee LF, et al. Elevated DDX21 regulates c-Jun activity and rRNA processing in human breast cancers. Breast Cancer Res 2014;16:449. [Crossref] [PubMed]
- Cao J, Wu N, Han Y, et al. DDX21 promotes gastric cancer proliferation by regulating cell cycle. Biochem Biophys Res Commun 2018;505:1189-94. [Crossref] [PubMed]
- Jung Y, Lee S, Choi HS, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res 2011;17:700-9. [Crossref] [PubMed]


