
SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis
Introduction
It is a well established scientific observation that mammalian cells contain fidelity or watchdog proteins that protect against the damaging effects of cellular stress (1). Thus, over the last 20 years a fundamental paradigm in biology has emerged proposing that these fidelity proteins recognize specific endogenous and exogenous forms of cell stress, including but not limited to oxidative and genotoxic stress and subsequently initiate signaling cascades that maintain cellular homeostasis (2,3). Loss of function or genetic mutation of specific fidelity proteins creates a cellular environment that is permissive for the development of genomic instability (4). According to the multi-hit genetic and epigenetic hypothesis for tumorigenesis, genomic instability is an early, initiating event, as it can trigger a phenotype favoring DNA damage and cellular transformation (5).
In this regard, loss of fidelity proteins has been shown to create a cellular environment permissive for genomic instability resulting in a cellular phenotype favoring DNA damage and cellular transformation (3). For example, it has been shown that mice lacking a fidelity protein, such as p53 and BRCA1, are susceptible to carcinogenesis and as such, these fidelity proteins are also referred to as tumor suppressor (TS) genes and/or proteins (3,6). In vitro it has also been shown that loss of function of a TS protein complements the activation of a single oncogene in the two-hit primary tissue culture cell model for carcinogenesis (7) and results in tumorigenesis in mice lacking expression (8). Critically, many common TS genes are deleted or mutated in human cancers, resulting in loss of function similar to knock-out mouse models (9,10). While these genes are often referred to as TS genes (TSGs) it seems unlikely that genetic evolution required proteins for the specific purpose to prevent tumors and it is more logical that these proteins regulate critical cell surveillance pathways. As such, it could be proposed that the subsequent loss of the aberrant function of critical cellular signaling pathways that protect the cell against damage from both endogenous as well as exogenous agents that induce stress may ultimately result in carcinogenesis.
One of the fundamental observations in oncology is that tumorigenesis increases as a function of age and in fact, increasing age is the strongest statistic variable that predicts for carcinogenesis (11,12). One fact that has emerged over the last several years is that aging is a complex cellular process that appears to be regulated, at least in part, by several signaling protein families that have been identified in multiple species, including a relatively new gene family that was initially identified in S. cerevisiae and C. elegans (12,13). The human and murine homologs of the yeast Sir2 gene are referred to as sirtuin proteins. While there is a controversy about the direct role of the sirtuin gene family in longevity in mammals they do appear to regulate critical signaling networks, and following stress, several mice lacking one of the sirtuin genes develop illnesses that mimic those observed in older humans (13,14).
Sirtuins are members of the class III histone deacetylase family of proteins and belong to the deoxyhypusine synthase (DHS)-like NAD/flavin adenine dinucleotide (FAD)-binding domain clan containing the Rossmann fold structural motif and phylogenetic analysis from a variety of prokaryotes and eukaryotes has divided the family into five different classes (15). In mammals, seven sirtuins (SIRT1-7) have been identified, each of them sharing a conserved 275-amino-acid catalytic core domain which are mainly categorized according to their subcellular localization to the nucleus (SIRT1, 6, and 7), mitochondria (SIRT3, 4, and 5), and cytoplasm (SIRT2), respectively (16). Besides histone deacetylation, lysine acetylation has recently emerged as an important, and perhaps a physiologically significant, post-translational modification employed to regulate several proteins (17-19). The reversible acetylation of lysine, which involves neutralization of a positive charge, alters protein structure and it seems very likely to also alter enzymatic function (20-23). Thus, it seems clear that sirtuins direct, at least in part, the cellular acetylome and may be fidelity or sensing proteins that respond to changes in the cellular environment and activate the catalytic to signaling activity of downstream target proteins to adapt to specific cellular conditions.
SIRT2 is the primary cytoplasmic sirtuin implicated in age related diseases
The SIRT2 protein is similar in sequence to yeast Hst2p and both proteins are located in the cytoplasm making it the first cytoplasmic sirtuin found (24). Consistent with the suggested role of sirtuins in the development of age-dependent disorders, SIRT2 has been found to regulate metabolism by deacetylating and stabilizing phosphoenolpyruvate carboxykinase (PEPCK1), which is the rate limiting enzyme for gluconeogenesis, linking SIRT2 with type II diabetes (25). In addition to its role in metabolic regulation, SIRT2 mediates the response to nutrient deprivation and energy expenditure by promoting lipolysis and inhibiting adipocyte differentiation through deacetylation of FoxO (25). Furthermore, p65, a subunit of NF-κB, was found to be another deacetylation target of SIRT2 in the cytoplasm where hyperacetylated p65 in Sirt2-/- cells after TNFa stimulation results in increased expression of a subset of NF-κB target genes implicated in the immune and inflammatory response (26). Finally, SIRT2 appears as an important regulator of neurodegeneration, however with opposite functional outcomes compared to other sirtuins. For example, whereas SIRT1 mainly exerts a neuroprotective role, SIRT2 seems to promote degeneration as revealed by studies showing that inhibition of SIRT2 rescues a-synuclein toxicity in a Parkinson’s disease model (27) and reduces mutant Huntington mediated toxicity caused by increased sterol levels (28), demonstrating a central function of SIRT2 in neurodegeneration.
Sirt2 functions as a murine tumor suppressor
As mentioned earlier, it is well documented that the incidence of malignant tumors increases progressively with age, in both animals and humans and cancer is one of the major life threatening age-related diseases. Thus, over the last years, the potential direct role of the sirtuins as longevity genes in tumorigenesis was a very interesting hypothesis that needed to be tested. Previous studies indicate that two members of the sirtuin family, Sirt1 and Sirt3, have tumor suppressor function (29-34). Based on these results it seemed reasonable to propose that the primary cytoplasmic sirtuin, Sirt2, might also function as a fidelity, watchdog, or TSG. In this regard the role of SIRT2 in tumorigenesis was relatively scarce and the results seemed somewhat conflicting. One example suggested that it might promote tumor formation based on the observation that SIRT2 deacetylates and inhibits the activity of p53 (35,36) and this might result in a tumor permissive phenotype. It was also shown that cell exposure to dual inhibitors of SIRT1 and SIRT2 induces apoptosis in tumor cell lines (36,37) and inhibits growth of Burkitt lymphoma xenografts (38), while an inhibitor for SIRT1 alone did not have such an anti-tumor effect (36). Furthermore a selective inhibitor of SIRT2 exhibited submicromolar selective cytotoxicity towards tumor cell lines compared to normal cell types by triggering apoptosis (37). Finally, a statistically stronger correlation between expression levels and pancreas tumor development was found in case of SIRT2 (6 of 11) compared to SIRT1 (1 of 11) (39).
In contrast, it was also published that SIRT2 expression is decreased in human gliomas, and ectopic expression of SIRT2 in glioma cell lines led to a remarkable reduction of in vitro colony formation ability (40). In addition, it has been suggested that loss of SIRT2 promotes genomic instability that is a well established early event in carcinogenesis (41-44). These results could be interpreted to suggest that SIRT2 functions as a fidelity protein or TSG. Consistently, in our recent study, we also showed that SIRT2 expression was decreased in genomic data in human breast cancer and hepatocellular carcinomas (HCCs), as compared to normal tissue human samples (45). Finally, analysis of the Oncomine cancer microarray database (46), at the University of Michigan, also revealed reduced SIRT2 mRNA expression in anaplastic oligodendroglioma, glioblastoma, clear cell renal carcinoma, and prostate carcinoma. Thus, these correlative results seemed to suggest a role of SIRT2 in carcinogenesis but the mechanism was unclear.
In order to further establish the role of Sirt2 as either a tumor suppressor or oncogene, our research groups analyzed the physiological function of SIRT2 in mutant mice generated by gene targeting. Experiments done using mouse embryo fibroblasts (MEFs) from these mice showed that Sirt2-deficient cells displayed centrosome amplification and several mitotic defects including failure to complete cytokinesis, arrest at metaphase and cell death during mitosis, resulting in genetic instability (45). Initially, Sirt2-deficient MEFs exhibited reduced proliferation; however, they gradually gained a more pro-proliferative phenotype and after multiple divisions became immortalized and finally displayed a series of malignant and transformed phenotypes as revealed by enhanced colony formation ability in vitro and tumor formation in vivo, implying that the absence of SIRT2 eventually triggers tumorigenesis. More importantly, it was also shown that the Sirt2-/- mice developed tumors in multiple tissues and the tumor incidence slowly increased with increasing mouse age (45) suggesting an essential role of Sirt2 in repressing tumor formation providing strong genetic evidence for a tumor suppressor function of a gene which is implicated with aging. So far, these recent results shift the balance to the tumor suppressor function of SIRT2 but also raise an intriguing question: why does the aberrant regulation of the Acetylome by SIRT2 result in a tumor permissive phenotype?
SIRT2 is a cytoplasmic sirtuin regulating mitosis
SIRT2 was originally reported as a cytoplasmic NAD dependent deacetylase that colocalizes with microtubules and specifically deacetylates lysine-40 of alpha-tubulin both in vitro and in vivo (47) establishing tubulin as the first bona fide substrate for SIRT2. However, it has also been reported that phosphorylation mediates the dramatic increase in the levels of the protein during the G2/M transition of the cell cycle (48), suggesting for the first time that SIRT2 may exert a key role in orchestrating mitotic events. In this regard, data showed that overexpression of the wild-type SIRT2 but not missense mutants lacking deacetylase activity results in a marked prolongation of the mitotic phase of the cell cycle (48). In addition, SIRT2 was found to deacetylate histone H4 lysine 16 (H4K16) mediating chromatin condensation during G2/M transition (49,50). These studies indicated that SIRT2 activity is required for mitotic progression and may be temporally and spatially controlled to aid in the faithful completion of mitosis. As a result, further studies focused on its role during the cell cycle and resulted in taking away the interest from its cytoplasmic localization. Further establishing its central role in mitosis, SIRT2 was found to be associated with mitotic structures, beginning with the centrosome during prophase, the mitotic spindle during metaphase, and the midbody during cytokinesis (51). Regarding its subcellular localization, it has been shown to accumulate in the nucleus in response to genotoxic stress such as ionizing radiation (52) while it is actively exported from the nucleus to maintain its cytoplasmic localization during interphase (51), indicating that nucleo-cytoplasm shuttling of SIRT2 is actively regulated in order to support the various cellular functions of the protein.
The role of SIRT2 as a gatekeeper of mitosis was further confirmed by data showing that overexpression of SIRT2 blocked the entry to chromosome condensation and subsequent hyperploid cell formation in glioma cells (52), whereas downregulation of SIRT2 prolonged chronic mitotic arrest from sustained activation of the mitotic checkpoint and consequently prevented a shift to secondary outcomes, including cell death, after release from chronic mitotic arrest rendering cancer cells resistant to microtubule inhibitors (53). While these observations link SIRT2 with cell division and thus maintenance of genomic stability, however more details remained to be elucidated regarding the mechanism of function and the identification of possible cell cycle specific proteins as new substrates for SIRT2 to explain the observed phenotypes during mitosis.
SIRT2 positively directs APC/C activity and prevents genomic instability
Since the observed abnormalities during cell cycle followed by widespread genetic instability in Sirt2-deficient cells could serve as a potential mechanism, at least in part, for the tumor permissive phenotype observed (45), we tried to shed more light on the underlying mechanism. For this purpose, we performed a proteomic study to identify interaction proteins and potential targets of SIRT2 deacetylase activity. After expressing a flag tagged SIRT2 in HeLa cells, followed by a pull-down using an antibody to flag, we identified several components of the APC/C complex, including CDH1 and CDC20 which serve as coactivators for APC/C and have substrate specificity for different APC/C substrates. In particular, we showed that CDH1 and CDC20 are downstream SIRT2 deacetylation targets and hyperacetylation reduces their interaction with CDC27 that decreases APC/C activity. In contrast, deacetylation of CDH1 and CDC20 by SIRT2 enhances their interaction with CDC27 leading to activation of APC/C suggesting that SIRT2 is a positive regulator for APC/C activity (45).
As such, it is proposed that the mitotic abnormalities can be caused by a combined effect of altered expression of mitotic regulators that are regulated by APC/C leading to perturbed maintenance of genomic integrity. For example, we indeed observed significantly higher levels of Aurora-A in Sirt2 mutant MEFs, mammary tissue and liver compared to wild-type control cells or tissues and previous studies have shown that overexpression of Aurora-A can cause centrosome amplification and mammary tumor formation in mice (54). Specifically, we found that knockdown of SIRT2 caused upregulation of Aurora-A which is associated with decreased ubiquitination, implying that E3 ubiquitin ligase activity of APC/C mediates this effect. Conversely, overexpression of the wild type SIRT2-WT, but not the deacetylase mutant SIRT2-HY, decreased the protein level of Aurora-A through increased ubiquitination. Finally, our analysis revealed increased levels of multiple mitotic regulators, including Aurora-A, Aurora-B, Plk1, securin, and cyclin A2 in Sirt2−/− cells, further supporting the key role of SIRT2 in regulating activity of APC/C complex (Figure 1).
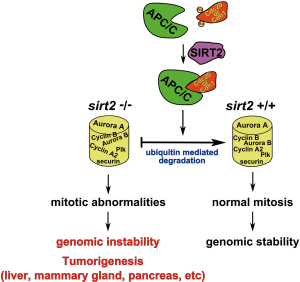
Since there are several lines of evidence suggesting that APC/C complex is involved in tumorigenesis (55,56), the interplay between SIRT2 and CDH1/CDC20 could provide the mechanistic link explaining how SIRT2 functions as a mitotic gatekeeper regulating the transition from metaphase to anaphase by controlling the proper formation of the complex through deacetylation of both CDC20 and CDH1 depending on the phase of the cell cycle. Thus, along with its well established localization with mitotic structures, SIRT2 is found to deacetylate targets that are directly involved in the proper completion of mitosis highlighting its role in maintaining genomic stability. Finally, the aberrant regulation of these factors, as well as others involved in cell segregation, may account for the tumor permissive phenotype, at least in part, in mice lacking Sirt2.
Conclusions and future aspects
In summary, Sirt2 is the third gene in the mammalian sirtuin gene family shown to be a tumor suppressor and the Sirt2-/- mice develop tumors as they age. In addition, SIRT2 directs mitotic events by regulating APC/C activity through deacetylation of its co-activators, CDH1 and CDC20. Thus, loss of Sirt2 consequently causes increased levels of many mitotic regulators that may contribute to centrosome amplification, aneuploidy, mitotic cell death, and most importantly the tumor permissive phenotype observed in the knockout mice. Finally, SIRT2 expression is reduced in several human malignancies including breast, liver, brain, kidney, and prostate cancers. Taken together, these results identify SIRT2 as a legitimate tumor suppressor gene and uncover an essential role for SIRT2 in maintaining the integrity of mitosis through positively regulating APC/C activity, a dysfunction of which leads to genetic instability and tumorigenesis. Although the central role of SIRT2 in regulating APC/C activity provides a link between SIRT2 and the observed mitotic defects reported in previous studies, however there are still questions that need to be answered regarding the specific role of the protein in tumorigenesis. For example, the gender specific profile of tumors developed in the Sirt2 knockout mice indicates that there are additional functions of SIRT2 that contribute possibly to tumor development in different tissues. Moreover even if genomic instability is an early event in tumorigenesis, it is clear that additional signaling pathways are required to be activated or deactivated in order to induce cell proliferation and it is not yet clear how SIRT2 regulates these pathways or how these changes are linked to the age-related cancer formation. Thus, further studies remain to uncover additional functions related to the role of SIRT2 as a tumor suppressor.
Acknowledgments
Melissa Stauffer, PhD, of Scientific Editing Solutions, provided editorial assistance.
Funding: DG is supported by 1R01CA152601-01 from the NCI, BC093803 from the DOD, SPORE P50CA98131, and a Hirshberg Foundation for Pancreatic Cancer Research Seed Grant Award. Dr. Deng’s laboratory was supported (in part) by the Intramural Research Program of the NIDDK, NCI, and CCR, NIH.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2012.05.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bishop JM. Viral oncogenes. Cell 1985;42:23-38. [PubMed]
- Gius D, Cui H, Bradbury CM, et al. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell 2004;6:361-71. [PubMed]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell 2002;2:103-12. [PubMed]
- Hunter T. Oncoprotein networks. Cell 1997;88:333-46. [PubMed]
- Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 1971;68:820-3. [PubMed]
- Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res 2006;34:1416-26. [PubMed]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 1983;304:596-602. [PubMed]
- Feinberg AP, Johnson LA, Law DJ, et al. Multiple tumor suppressor genes in multistep carcinogenesis. Tohoku J Exp Med 1992;168:149-52. [PubMed]
- Land H, Chen AC, Morgenstern JP, et al. Behavior of myc and ras oncogenes in transformation of rat embryo fibroblasts. Mol Cell Biol 1986;6:1917-25. [PubMed]
- Sherr CJ. Principles of tumor suppression. Cell 2004;116:235-46. [PubMed]
- Ershler WB, Longo DL. The biology of aging: the current research agenda. Cancer 1997;80:1284-93. [PubMed]
- Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell 2006;126:257-68. [PubMed]
- Kim EJ, Kho JH, Kang MR, et al. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell 2007;28:277-90. [PubMed]
- Wood JG, Rogina B, Lavu S, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 2004;430:686-9. [PubMed]
- Vassilopoulos A, Fritz KS, Petersen DR, et al. The human sirtuin family: evolutionary divergences and functions. Hum Genomics 2011;5:485-96. [PubMed]
- Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3?--functional localization of an NAD+-dependent protein deacetylase. Biochem J 2008;411:e11-e13. [PubMed]
- Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 2008;31:449-61. [PubMed]
- Schwer B, Bunkenborg J, Verdin RO, et al. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA 2006;103:10224-9. [PubMed]
- Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009;325:834-40. [PubMed]
- Kouzarides T. Chromatin modifications and their function. Cell 2007;128:693-705. [PubMed]
- Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 2004;26:1076-87. [PubMed]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 2007;76:75-100. [PubMed]
- Yang H, Baur JA, Chen A, et al. Design and synthesis of compounds that extend yeast replicative lifespan. Aging Cell 2007;6:35-43. [PubMed]
- Afshar G, Murnane JP. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene 1999;234:161-8. [PubMed]
- Jiang W, Wang S, Xiao M, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell 2011;43:33-44. [PubMed]
- Rothgiesser KM, Erener S, Waibel S, et al. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci 2010;123:4251-8. [PubMed]
- Outeiro TF, Kontopoulos E, Altmann SM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 2007;317:516-9. [PubMed]
- Luthi-Carter R, Taylor DM, Pallos J, et al. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc Natl Acad Sci USA 2010;107:7927-32. [PubMed]
- Wang RH, Sengupta K, Li C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 2008;14:312-23. [PubMed]
- Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci 2009;5:147-52. [PubMed]
- Kim HS, Patel K, Muldoon-Jacobs K, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010;17:41-52. [PubMed]
- Tao R, Coleman MC, Pennington JD, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 2010;40:893-904. [PubMed]
- Bell EL, Emerling BM, Ricoult SJ, et al. SirT3 suppresses hypoxia inducible factor 1 and tumor growth by inhibiting mitochondrial ROS production. Oncogene 2011;30:2986-96. [PubMed]
- Finley LW, Carracedo A, Lee J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1 destabilization. Cancer Cell 2011;19:416-28. [PubMed]
- Jin YH, Kim YJ, Kim DW, et al. Sirt2 interacts with 14-3-3 beta/gamma and down-regulates the activity of p53. Biochem Biophys Res Commun 2008;368:690-5. [PubMed]
- Peck B, Chen CY, Ho KK, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther 2010;9:844-55. [PubMed]
- Zhang Y, Au Q, Zhang M, et al. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem Biophys Res Commun 2009;386:729-33. [PubMed]
- Heltweg B, Gatbonton T, Schuler AD, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res 2006;66:4368-77. [PubMed]
- Ouaïssi M, Sielezneff I, Silvestre R, et al. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol 2008;15:2318-28. [PubMed]
- Hiratsuka M, Inoue T, Toda T, et al. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun 2003;309:558-66. [PubMed]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 2000;14:1021-6. [PubMed]
- Kyrylenko S, Kyrylenko O, Suuronen T, et al. Differential regulation of the Sir2 histone deacetylase gene family by inhibitors of class I and II histone deacetylases. Cellular and molecular life sciences. Cell Mol Life Sci 2003;60:1990-7. [PubMed]
- Lombard DB, Chua KF, Mostoslavsky R, et al. DNA repair, genome stability, and aging. Cell 2005;120:497-512. [PubMed]
- Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006;124:315-29. [PubMed]
- Kim HS, Vassilopoulos A, Wang RH, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 2011;20:487-99. [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [PubMed]
- North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol 2004;5:224. [PubMed]
- Dryden SC, Nahhas FA, Nowak JE, et al. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol 2003;23:3173-85. [PubMed]
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 2007;26:5505-20. [PubMed]
- Vaquero A, Scher MB, Lee DH, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 2006;20:1256-61. [PubMed]
- North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS ONE 2007;2:e784 [PubMed]
- Inoue T, Hiratsuka M, Osaki M, et al. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene 2007;26:945-7. [PubMed]
- Inoue T, Nakayama Y, Yamada H, et al. SIRT2 downregulation confers resistance to microtubule inhibitors by prolonging chronic mitotic arrest. Cell Cycle 2009;8:1279-91. [PubMed]
- Wang X, Zhou YX, Qiao W, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene 2006;25:7148-58. [PubMed]
- Saeki K, Iwasa Y. Optimal number of regulatory T cells. J Theor Biol 2010;263:210-8. [PubMed]
- García-Higuera I, Manchado E, Dubus P, et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol 2008;10:802-11. [PubMed]


