
Gene expression profile of THZ1-treated nasopharyngeal carcinoma cell lines indicates its involvement in the inhibition of the cell cycle
Introduction
Nasopharyngeal carcinoma (NPC) is a common type of head and neck tumor. According to global cancer statistics, approximately 86,700 new NPC cases and 50,800 deaths occurred worldwide in 2012 (1). Approximately 92% of new diagnoses occur in developing countries, and the incidence of NPC is two to threefold higher in men than in women (1). In recent years, NPC cases account for 38.29% and 40.14% of the global NPC morbidity and mortality, respectively (2). Guangdong and Guangxi have been identified as high-incidence areas for this type of cancer (2). With the increase in the aging population, smoking, air pollution, Epstein–Barr virus (EBV) infection, and other risk factors, the cancer burden caused by NPC will increase in the future, thereby posing a threat to public health, particularly in epidemic areas. Nasopharyngectomy is one of the effective treatments of NPC, however, is more difficult and can lead to substantial morbidity. Currently, radiotherapy is the first choice for early NPC treatment, and the combination of radiotherapy and chemotherapy are used as treatment for locally advanced NPC (3). However, for poorly tolerated patients, the effect of radiotherapy may not be ideal. It is indicated that immunotherapy, such as anti-PD-1 antibody nivolumab, is an effective treatment on advanced patients through the blockade of immune check-point (4,5). Therefore, it is necessary to find an accurate predictive marker.
THZ1 is a covalent cyclin-dependent kinase 7 (CDK7) inhibitor that targets a remote cysteine residue located outside the canonical kinase domain (6). THZ1 bears a potentially cysteine-reactive acrylamide moiety and shows to suppress cell proliferation and biochemical CDK7 activity (6). In Jurkat cells, THZ1 treatment had a profound effect on the downstream expression of some genes, particularly RUNX1, thereby contributing to the subsequent loss of the gene expression program and cell death (6). THZ1 is currently emerging as a novel cancer targeted pharmacological strategies (7), including peripheral blood T-cell lymphoma (8), breast cancer (9,10), non-small cell lung cancer (11), pancreatic ductal adenocarcinoma (12) and neuroblastoma (13).
RNA-seq is a quick turnaround high-throughput sequencing technology that has recently become an effective tool for the analysis of gene expression profiles. Functional enrichment analysis and protein–protein interaction (PPI) network construction provide tools to identify novel genes and pathways in THZ1-sensitive NPC. Current studies have identified potential biomarkers of NPC. Such as the microarray seq study shows that miR-135b, miR-574-5p, miR-BART7-3p and miR-BART13-3p have a good diagnosis value of NPC (14,15). CircRNA sequencing result shows that hsa_circ_001387 and hsa_circ_0002375 can be used as biomarkers for NPC (16,17). DNA methylation seq finds a variety of DEGs related to NPC, among which FANCI, POSTN, IFIH1, ZMYND10, PACRG and POU2AF1 can be used as new biomarkers for NPC patients (18,19).
In this study, we acquired four whole transcriptome sequencing datasets for NPC (untreated or THZ1-treated) from an integrative research of the literature. After analysis of differentially expressed genes (DEGs), Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, and PPI construction, we identified several DEGs, novel KEGG pathways and node genes associated with THZ1 sensitivity in NPC. These results may be further exploited in the future for the development of more effective therapeutics for NPC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-19-2888).
Methods
Data preparation of transcriptome
Four gene expression datasets were collected from the National Center for Biotechnology Information Gene Expression Omnibus (GEO), included GSM2523140 (C666-1 cell line treated with DMSO), GSM2523141 (C666-1 cell line treated with THZ1), GSM2523142 (HK1 cell line treated with DMSO), and GSM2523143 (HK1 cell line treated with THZ1). RNA-seq was performed using the Illumina HiSeq 2000 platform.
Analysis and identification of DEGs
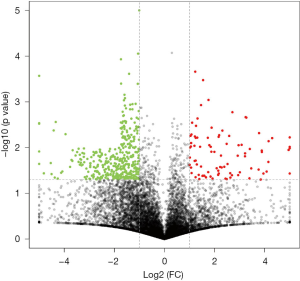
Gene expression datasets before and after THZ1 treatment were obtained from the GEO datasets (GSE95750). The corresponding gene symbols of the probe sets were obtained through the annotation package, and gene expression was summarized. A P value <0.05 and log (fold change) >2 were used to screen for DEGs. The volcano plot and heat-map of gene expression data were constructed using R-language (ggplot 2 package).
Cell culture and treatment of THZ1
Human NPC cell line C666-1 cell line and HK1 cell line were obtained from Shanghai Guandao Biological Engineering Co., Ltd. [#GDC (242349382-03), Shanghai, China] and Shanghai Meiyan Biological Engineering Co., Ltd (# ATCC-1435, Shanghai, China) respectively. C666-1 cell line was grown in 1640 medium and HK1 cell line was cultured in Dulbecco’s modification of Eagle’s medium. C666-1 cell line and HK-1 cell line were then treated with THZ1 (ApexBio, Houston, TX, USA) at concentration of 200 nM for 6 h, named as THZ1-treated group, and treated by THZ1 solvent dimethylsulfoxide (DMSO) named as untreated group.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis for DEGs verification
Total RNA was extracted from both C666-1 and HK1 cells using an RNA Easy Fast extraction kit (Tiangen, Beijing, China) and then reverse transcribed into cDNA using PrimeScript RT Reagent Kit (Qiagen, Valencia, CA, USA). PCR was performed with primers (Table 1) using a miScript SYBR Green PCR kit (Qiagen). 2–ΔΔCt method was used for quantifying the expression of each gene. The relative expression of the detected genes was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Experiments were performed in triplicate.
Table 1
| Primer name | Sequences (5'-3') |
|---|---|
| AC010970.2-F | ATAATTGCAATCCCCGATCA |
| AC010970.2-R | TTTAGCCACCCGAGATTGAG |
| GPX1P1-F | GCACACTCTCTTTGCCTTCC |
| GPX1P1-R | CTCGATGTCAATGGTCTGGA |
| CHRAC1-F | ACGTGGTCGTGGGTAAAGAC |
| CHRAC1-R | TCCTGGTTGATGCTGGACAC |
| HERC3-F | AAGGTGTGTGGTGGCAAAGT |
| HERC3-R | GGGATGTGTGGCCGAGTAAT |
| FAM89A-F | AAGAGATGGTTGGTCTCCGC |
| FAM89A-R | TCCAGAGCGTAAGTGCAGTC |
| PPP1R10-F | AACCAGCACCACCTTCTGAG |
| PPP1R10-R | TCCGGGTCAGTTGGTTAGGA |
| PNRC2-F | CCTGGCAGGCCATGCAAAAT |
| PNRC2-R | GAAGAACACTTGGTGATGGCG |
| TRIAP1-F | ACATGAAGCGCGAGTACGAC |
| TRIAP1-R | TGAACACACTGCTGGTAGCG |
| NXT1-F | ATGTTGCCTTCCAGCGAGTT |
| NXT1-R | GTCCCGTTGTTTGTTCCCCT |
| DDIT4-F | TGAACACTTGTGTGCCAACC |
| DDIT4-R | CCAGGCGCAGTAGTTCTTTG |
| CDC6-F | GCAGTTCAATTCTGTGCCCG |
| CDC6-R | TAGCTCTCCTGCAAACATCCAG |
| CDC34-F | AACGAGCCCAACACCTTCTC |
| CDC34-R | ACCTGCTTCCGGATGATGTC |
| CDK7-F | GGTCTCCTTGATGCTTTTGGAC |
| CDK7-R | GTGTGATGGTGTCAGCACAAG |
| CDK9-F | CATTACAGCCTTGCGGGAGA |
| CDK9-R | ACCCTTGCAGCGGTTATAGG |
| CCNA2-F | GGTACTGAAGTCCGGGAACC |
| CCNA2-R | CATGAATGGTGAACGCAGGC |
| CCNB1-F | GCACTTCCTTCGGAGAGCAT |
| CCNB1-R | TGTTCTTGACAGTCCATTCACCA |
| CDT1-F | ATGCGTAGGCGTTTTGAGGA |
| CDT1-R | GCTCGATGGTGAGCTGGTAA |
| KIF11-F | TGGCTGACAAGAGCTCAAGG |
| KIF11-R | GGCCATACGCAAAGATAGTGC |
| LIN9-F | GACCAGTTGCCTGACGAGAG |
| LIN9-R | TCCTGCCTTTCCAAACAGGT |
| PLK1-F | TGACTCAACACGCCTCATCC |
| PLK1-R | GCTCGCTCATGTAATTGCGG |
| POLR3E-F | TGACATTCCGCACCTCTCAG |
| POLR3E-R | GGAATACGTGCTGGTCTCGT |
| POLR3C-F | CTGATAAGAACCGGCAGCCA |
| POLR3C-R | ACACCACGTTTGTGCACTTG |
| POLR1B-F | GGGAACAGCAAAAGGCAGC |
| POLR1B-R | TTGATATCAGCCTGCACCGC |
| POLR1E-F | TTCTGGAAAGAGCCAAGGACT |
| POLR1E-R | AGCGCCAGTTCACTCTCAAC |
| POLR3A-F | GAGACGGATGTGGCCAAGAA |
| POLR3A-R | CATCCTATGGTCGAGCACCC |
| GAPDH-F | GACCACAGTCCATGCCATCA |
| GAPDH-R | CCGTTCAGCTCAGGGATGAC |
GO enrichment analysis
GO enrichment analysis of DEGs was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). The functions of DEGs was analyzed and enriched into biological processes (BP), cellular components (CC), and molecular functions (MF). And P value <0.05 was defined as the cutoff criterion for significance.
KEGG pathway analysis
KEGG enrichment analysis was conducted for the selected drug-sensitive genes and pathways using DAVID. The pathways in untreated group and THZ1-treated group was investigated. And P value <0.05 was considered as statistically significant.
PPI network construction and gene-interaction network
PPI analysis is essential to interpret the molecular mechanisms of key cellular pathways in NPC. In this study, the Search Tool in the Retrieval of Interacting Genes database was used to construct a PPI network with the 592 THZ1-sensitive genes. An interaction score of 0.4 was considered as the cutoff criterion. The Cytoscape software was used to screen for significant modules within the PPI network.
Statistical analysis
Data was from at three biological replicates, presented as mean ± standard deviation, which was analyzed using the SPSS 17.0 software. The nonparametric student t-test was performed to define significant differences between two group, using a P value of <0.05.
Results
Identification of DEGs in THZ1-treated NPC cell lines
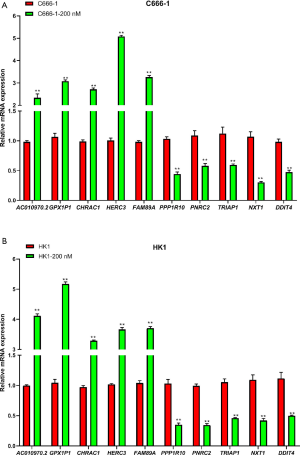
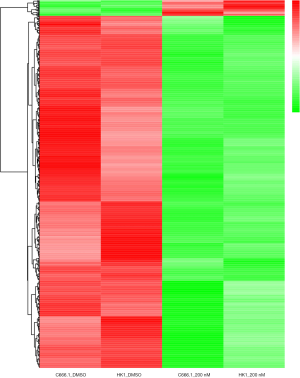
Data from each cell line were separately analyzed to screen for DEGs, and 25 upregulated genes and 567 downregulated genes were subsequently identified. The chromosome distribution of DEGs is shown in Figure 1. The top 10 most significant DEGs are listed in Table 2, with further details shown in in total online: + https://cdn.amegroups.cn/static/public/10.21037tcr-19-2888-1.xlsx. The five most significantly upregulated DEGs between untreated group and THZ1-treated group were AC010970.2, GPX1P1, CHRAC1, HERC3, and FAM89A, and the five most significantly downregulated DEGs were PPP1R10, PNRC2, TRIAP1, NXT1, and DDIT4. The in vitro validation experiment shown that the expression of all five upregulated and five downregulated DEGs in THZ1-treated C666 and HK1 cell lines verified by qRT-PCR were in agreement with the results of RNA-seq (Figure 2). The representative heat-map and volcano plots for GSE95750 are shown in Figures 3,4.
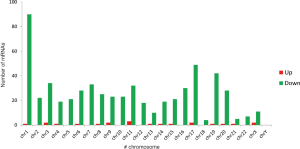
Table 2
| Gene name | C666-1_DMSO | HK1_DMSO | C666-1_200nM | HK1_200nM | Fold change | P value |
|---|---|---|---|---|---|---|
| TOP 5 up-regulated genes | ||||||
| AC010970.2 | 4.621964 | 5.28916 | 266.3011 | 394.9896 | 66.72207 | 0.036886 |
| GPX1P1 | 2.870082 | 0.919901 | 37.60978 | 26.4766 | 16.90941 | 0.033389 |
| CHRAC1 | 4.656576 | 3.089185 | 18.01039 | 16.66689 | 4.476936 | 0.005824 |
| HERC3 | 3.563076 | 2.592196 | 9.778549 | 13.53734 | 3.787955 | 0.047557 |
| FAM89A | 1.718694 | 1.088639 | 4.947675 | 4.771074 | 3.461915 | 0.008845 |
| TOP 5 down-regulated genes | ||||||
| PPP1R10 | 26.72145 | 22.09977 | 0.997374 | 1.939122 | 0.060148 | 0.010402 |
| PNRC2 | 32.35008 | 34.78629 | 2.085106 | 1.359871 | 0.051313 | 0.001589 |
| TRIAP1 | 38.97422 | 25.05203 | 1.556227 | 1.243586 | 0.043729 | 0.048035 |
| NXT1 | 18.22963 | 23.25776 | 0.516605 | 1.243586 | 0.041244 | 0.015883 |
| DDIT4 | 38.20736 | 56.01591 | 0.946979 | 2.407141 | 0.035598 | 0.036559 |
NPC, nasopharyngeal carcinoma.
GO and KEGG pathways associated with drug-specific sensitivity
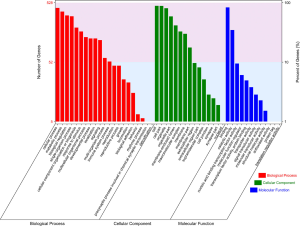
The significant terms for the GO enrichment analysis in DAVID are illustrated in Figure 5. Most DEGs were enriched for the BP of cellular process, metabolic process, biological regulation, single-organism process, and CC organization or biogenesis (top five terms). Regarding MF, DEGs showed enrichment in binding, catalytic activity, nucleic acid binding transcription factor activity, MF regulator, and transcription factor activity and protein binding (top five terms). Regarding CC, enrichment was predominantly within the cell, cell part, organelle, organelle part, and membrane-enclosed lumen terms (top five terms). These enrichment terms indicate that these DEGs may play a crucial role in NPC tumorigenesis. DEGs enrichment terms are shown in in total online: + https://cdn.amegroups.cn/static/public/10.21037tcr-19-2888-2.xls.
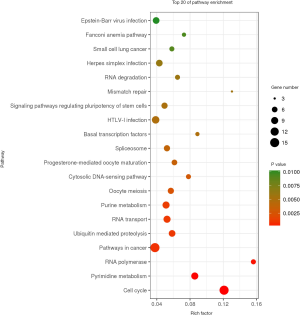
KEGG pathway enrichment analysis was performed in DAVID. A total of 592 DEGs were found enriched in 38 KEGG pathways, including cell cycle, pathways in cancer, HTLV-I infection, Herpes simplex infection, and EBV infection (Figure 6, in total online: + https://cdn.amegroups.cn/static/public/10.21037tcr-19-2888-3.xls).
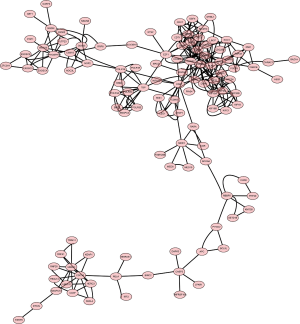
PPI network and novel node genes associated with drug-specific sensitivity
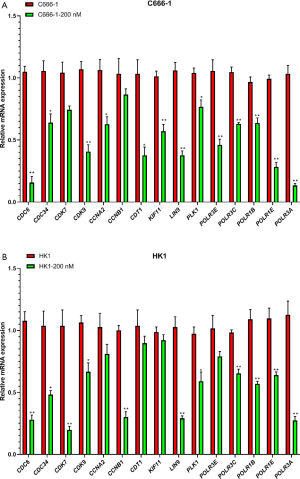
RNA-seq analysis reveals five networks of molecular associations for the DEGs in untreated group versus THZ1-treated group. The PPI network complex contains 176 nodes and 366 edges. All 592 DEGs are involved in these networks. The most significant network connection is related to the cell cycle. We identified several clusters in this sub-module using the MCODE plugin, including CDC6, CDC34, CDK7, CDK9, CCNA2, CCNB1, CDT1, KIF11, LIN9, PLK1, and POLR family (Figure 7). qRT-PCR was used to validate the expression of predicted clusters. And the results show that the expression of all clusters are downregulated in THZ1-treated C666-1 and HK1 compare to that untreated group, which are consistent with the results of RNA-seq (Figure 8).
Discussion
In this study, we identified 25 up-regulated genes and 567 down-regulated genes between THZ1-treated NPC cells and normal NPC cell lines (treated with DMSO). The functional analysis including GO terms and KEGG pathways indicated the most important enriched pathways were cell cycle. PPI network complex further identified several related node genes, such as CDC6, CDC34, CDK7, CDK9, CCNA2, CCNB1, CDT1, KIF11, LIN9, PLK1, and POLR family. Our study indicated that THZ1 may be potential effective therapeutics for NPC and provided new sight for investigating the pharmacological mechanism of THZ1.
NPC is one of the most common malignant tumors in South China and Southeast Asia (20). It occurs on the top and side wall of the nasopharyngeal cavity (20). The pathogenesis of NPC is still unclear. In addition to genetic factors, it is also closely related to the environment, living habits, and other factors. A World Health Organization survey reported that 80% of NPC cases occur in China (9). In recent years, the incidence of NPC in China has increased gradually. With the continuous improvement in the diagnosis and treatment of NPC, the curative effects of advanced NPC have also improved. However, the 5-year survival rate of patients is still 70% (9). Molecular targeted drugs for cell signaling pathways have guided the new era of tumor therapy, and it is important to explore effective targets for tumor diagnosis and treatment. In this study, 25 drug-sensitive genes were found upregulated and 567 downregulated that were identified through data analysis of GSE95754 in NPC using multiple bioinformatics tools. Enrichment analysis of the identified DEGs demonstrated gene nodes and pathways sensitive to THZ1, providing new insights for the diagnosis and therapy of NPC.
In this study, we identified the cell cycle as the most affected pathway by THZ1. The cell cycle is one of the most important functions in cells, and an abnormal cell cycle is closely associated with diseases such as cancer. The cell cycle drives cells’ proliferation by chromosome duplication and distribution to daughter cells. These processes rely on a series of sequential steps that control the transition of cells from the S phase to the M phase, separated by gap phases G1 and G2 (21). The activation of CDKs and association with regulatory cyclins regulate the successful progression through the cell cycle (22). Nine CDKs form cyclin/CDK complexes that regulate different processes during cell cycle progression (22). In our study, we identified a series of genes that play crucial roles in each step of the cell cycle, including CDC6, CDC34, CDK7, CDK9, CCNA2, CCNB1, CDT1, KIF11, LIN9, PLK1, and the POLR gene family. These were downregulated in THZ1-treated NPC cell lines compared with untreated cells.
The process of DNA replication is tightly regulated by a protein complex consisting of CDC6, CDT1, origin-recognition complex (ORC), and minichromosome maintenance proteins (MCMs) (23). During the period starting in the late M phase to the early G1 phase, ORC binds to the DNA replication origin and recruits CDC6 and CDT1 (24). CDC6 is an androgen receptor target gene implicated in DNA replication and checkpoint mechanisms. The best characterized function of CDC6 is the pre-replicative complex assembly at the origin of replication (25). Overexpression of CDC6 during the G2 phase suppresses the mitotic entry by activating CHK1, which in turn blocks G2/M progression (26,27). CDC6 interacts with ATR and promotes the activation of a replication checkpoint (27). Genomic instability decreased by CDC6 is vital for cancer cell survival. CDC6 can regulate oncogenic activity via the regulation of DNA replication and repression of tumor suppressors (28). Increased CDC6 expression was also found to suppress p14, p15, p16, and E-cadherin (29).
CDT1 is another key regulatory factor in DNA replication, and the cellular level of CDT1 is critically regulated by ubiquitin-mediated degradation and inhibited by geminin (30). High levels of CDT1 occur during mitosis and in the G1 phase and low levels during the S and G2 phases (31-33). Deletion of either CDC6 or CDT1 suppresses the normal association of MCMs with chromatin during the G1 phase (34). In contrast, overexpression of CDC6 and CDT1 contributes to tumorigenesis and is associated with cancer progression in various types of cancers (35-39). Targeting the CDC6-ATR-CHK1 signaling pathway increases the sensitivity to treatment with agents, making this method particularly attractive for cancer therapy (40).
CDC34 is an E2 ubiquitin-conjugating enzyme that controls the ubiquitin-dependent proteolysis during cell cycle progression (41). CDC34 affects the ubiquitin protein function, which leads to the degradation of CDC6 (42). CDC34 can also control the degradation of p27KIP1 to regulate the cell cycle at the G1 phase (43). Abnormal overexpression of CDC34 has been found in hepatocellular carcinomas, and the non-coding RNA let-7d/CDC34 axis contributed to niclosamide-induced cell cycle arrest in G1 in head and neck squamous cell carcinoma (44,45). This may indicate that the CDC family, particularly CDC6 and CDC34, participate in the THZ1-induced inhibition of NPC cell proliferation.
Another gene family that we found associated with THZ1 drug sensitivity was the CDK family. CDKs are an enzyme family that associates with the cyclin family (46). They work together to form functional heterodimeric complexes responsible for several cellular functions, including cell cycle, differentiation, DNA repair, and apoptosis. CDKs are proline-directed serine/threonine protein kinases. Their characterized structural and molecular mechanisms have become attractive pharmacological targets for the development of antiproliferative drugs (46). The significant differential expression of CDK genes included both CDK7 and CDK9. CDK7 and CDK9 are located in the nodal position in the PPI network analysis. CDK7 is a major member of the CDK family that together with cyclin H and MAT1 forms a CDK-activating kinase that participates in the cell cycle by phosphorylating other CDKs (47)(48). CDK7 is also a component of transcription factor II H, which regulates transcriptional initiation and elongation (49-51). It has been previously demonstrated that the inhibition of CDK7 function may provide an effective therapeutic method for the treatment of cervical cancer by suppressing cell cycle progression and transcriptional activity. THZ1 is a dominant inhibitor of CDK7 and can inhibit the cell cycle and repress essential oncogene transcription during tumorigenesis (52). THZ1-induced CDK7 inhibition downregulated cyclin B1, a cell cycle checkpoint mediator, and CDK1 phosphorylation, inducing cell cycle arrest at the G2/M phase (52). CDK9 is located on chromosome 9q34.1 and is a main factor in RNA polymerase II transcription regulation (53). Aberrant CDK9 expression occurs in numerous types of cancers and recruits positive activation of factor b, which is essential for the gene transcription of MYC. MYC is a proto-oncogene located downstream of CDK9 that is known to be involved in cell cycle and cell growth (54). Over-stimulation of CDK9 is one of the proposed leading mechanisms of cancer development (55). Decreased expression of CDK9 leads to suppression of p53 protein activity and allows for DNA damaged cells to continue dividing (56). Collectively, these observations indicate that cell cycle networks, including CDK6 and CDK9, are associated with THZ1-induced anti-tumor effects.
Cyclin A2 (CCNA2) is a cell cycle regulatory factor that contributes to cell cycle procession and plays important roles in cell proliferation (57). CCNA2 is critical for the transition of the G1/S and the G2/M and its expression is essential in embryonic cells and in the hematopoietic lineage (58). CCNA2 has been found to be overexpressed in several types of cancer, which indicates its crucial role in cancer transformation and progression (59). CCNA2 was also shown to be involved in the process of epithelial-to-mesenchymal transition and in metastasis (60). CCNB1 is another member of the cyclin family and a key initiator of mitosis. CCNB1 can form a complex with CDK1, which in turns phosphorylates its substrates to promote the transition from the G2 phase to mitosis (61,62). CCNB1 also has a demonstrated role in checkpoint control. A dysfunction in CCNB1 is an early event in cancer, and aberrant CCNB1 expression is observed in various types of cancer (63,64). CCNB1 was also considered as an independent predictor of HBV-positive hepatocellular carcinoma (65). Aberrant CCNA2 and CCNB1 expression implies that these genes might contribute to the anti-tumor effects of THZ1. In addition, other node genes, such as KIF11, LIN9, PLK1, and POLR family, may correlate with THZ1-induced inhibition of cell proliferation. However, evidence for these is currently scarce.
Conclusions
In summary, we identified several novel node genes and pathways regulated by THZ1 in NPC. These data provide a basis for the understanding of the anti-tumor mechanisms induced by THZ1.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at: http://dx.doi.org/10.21037/tcr-19-2888
Data Sharing Statement: Available at: http://dx.doi.org/10.21037/tcr-19-2888
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2888). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Xin L, Jian Y, Ting G, et al. Nasopharynx Cancer Epidemiology in China. China Cancer 2016;25:835-40.
- Holliday EB, Frank SJ. Proton therapy for nasopharyngeal carcinoma. Chin Clin Oncol 2016;5:25. [Crossref] [PubMed]
- Chen YP, Tang LL, Yang Q, et al. Induction chemotherapy plus concurrent chemoradiotherapy in endemic nasopharyngeal carcinoma: Individual patient data pooled analysis of four randomized trials. Clin Cancer Res 2018;24:1824-33. [Crossref] [PubMed]
- Ma BBY, Lim WT, Goh BC, et al. Antitumor activity of nivolumab in recurrent and metastatic nasopharyngeal carcinoma: An international, multicenter study of the mayo clinic phase 2 consortium (NCI-9742). J Clin Oncol 2018;36:1412-8. [Crossref] [PubMed]
- Kwiatkowski N, Zhang T, Rahl PB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014;511:616-20. [Crossref] [PubMed]
- Greenall SA, Lim YC, Mitchell CB, et al. Cyclin-dependent kinase 7 is a therapeutic target in high-grade glioma. Oncogenesis 2017;6:e336. [Crossref] [PubMed]
- Cayrol F, Praditsuktavorn P, Fernando TM, et al. THZ1 targeting CDK7 suppresses STAT transcriptional activity and sensitizes T-cell lymphomas to BCL2 inhibitors. Nat Commun 2017;8:14290. [Crossref] [PubMed]
- Tang L, Jin J, Xu K, et al. SOX9 interacts with FOXC1 to activate MYC and regulate CDK7 inhibitor sensitivity in triple-negative breast cancer. Oncogenesis 2020;9:47. [Crossref] [PubMed]
- McDermott MSJ, Sharko AC, Munie J, et al. CDK7 Inhibition Is Effective in all the Subtypes of Breast Cancer: Determinants of Response and Synergy with EGFR Inhibition. Cells 2020;9:638. [Crossref] [PubMed]
- Wang J, Zhang R, Lin Z, et al. CDK7 inhibitor THZ1 enhances antiPD-1 therapy efficacy via the p38α/MYC/PD-L1 signaling in non-small cell lung cancer. J Hematol Oncol 2020;13:99. [Crossref] [PubMed]
- Huang CS, You X, Dai C, et al. Targeting Super-Enhancers via Nanoparticle-Facilitated BRD4 and CDK7 Inhibitors Synergistically Suppresses Pancreatic Ductal Adenocarcinoma. Adv Sci (Weinh) 2020;7:1902926. [Crossref] [PubMed]
- Tee AE, Ciampa OC, Wong M, et al. Combination therapy with the CDK7 inhibitor and the tyrosine kinase inhibitor exerts synergistic anticancer effects against MYCN-amplified neuroblastoma. Int J Cancer 2020;147:1928-38. [Crossref] [PubMed]
- Zhang G, Zong J, Lin S, et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer 2015;136:E301-12. [Crossref] [PubMed]
- Mi JL, Xu M, Liu C, et al. Identification of novel biomarkers and small-molecule compounds for nasopharyngeal carcinoma with metastasis. Medicine (Baltimore) 2020;99:e21505. [Crossref] [PubMed]
- Shuai M, Huang L. High expression of hsa_circrna_001387 in nasopharyngeal carcinoma and the effect on efficacy of radiotherapy. Onco Targets Ther 2020;13:3965-73. [Crossref] [PubMed]
- Yang J, Gong Y, Jiang Q, et al. circular RNA expression profiles in nasopharyngeal carcinoma by sequencing analysis. Front Oncol 2020;10:1-12.
- Wu ZH, Zhou T, Sun HY. DNA methylation-based diagnostic and prognostic biomarkers of nasopharyngeal carcinoma patients. Medicine (Baltimore) 2020;99:e20682. [Crossref] [PubMed]
- Han B, Yang X, Zhang P, et al. DNA methylation biomarkers for nasopharyngeal carcinoma. PLoS One 2020;15:e0230524. [Crossref] [PubMed]
- Wei KR, Zheng RS, Zhang SW, et al. Nasopharyngeal carcinoma incidence and mortality in China in 2010. Chin J Cancer 2014;33:381-7. [PubMed]
- Mills CC, Kolb EA, Sampson VB. Recent advances of cell-cycle inhibitor therapies for pediatric cancer. Cancer Res 2017;77:6489-98. [Crossref] [PubMed]
- Malumbres M. Cyclin-dependent kinases. Genome Biol 2014;15:122. [Crossref] [PubMed]
- Mahadevappa R, Neves H, Yuen SM, et al. The prognostic significance of Cdc6 and Cdt1 in breast cancer /692/4028/67/1347 /692/4028/67/1857 /13 /13/109 /38 /13/106 /38/39 /82/80 article. Sci Rep 2017;7:985. [Crossref] [PubMed]
- Wu L, Liu Y, Kong DC. Mechanism of chromosomal DNA replication initiation and replication fork stabilization in eukaryotes. Sci China Life Sci 2014;57:482-7. [Crossref] [PubMed]
- Donzelli M, Draetta GF. Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep 2003;4:671-7. [Crossref] [PubMed]
- Clay-Farrace L, Pelizon C, Santamaria D, et al. Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. EMBO J 2003;22:704-12. [Crossref] [PubMed]
- Yoshida K, Sugimoto N, Iwahori S, et al. CDC6 interaction with ATR regulates activation of a replication checkpoint in higher eukaryotic cells. J Cell Sci 2010;123:225-35. [Crossref] [PubMed]
- Gonzalez S, Klatt P, Delgado S, et al. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature 2006;440:702-6. [Crossref] [PubMed]
- Sideridou M, Zakopoulou R, Evangelou K, et al. Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins. J Cell Biol 2011;195:1123-40. [Crossref] [PubMed]
- Saxena S, Dutta A. Geminin-Cdt1 balance is critical for genetic stability. Mutat Res 2005;569:111-21. [Crossref] [PubMed]
- Nishitani H, Taraviras S, Lygerou Z, et al. The Human Licensing Factor for DNA Replication Cdt1 Accumulates in G 1 and Is Destabilized after Initiation of S-phase. J Biol Chem 2001;276:44905-11. [Crossref] [PubMed]
- Rialland M, Sola F, Santocanale C. Essential role of human CDT1 in DNA replication and chromatin licensing. J Cell Sci 2002;115:1435-40. [PubMed]
- Wohlschlegel JA, Dwyer BT, Dhar SK, et al. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science (80-) 2000;290:2309-12.
- Cook JG, Chasse DAD, Nevins JR. The Regulated Association of Cdt1 with Minichromosome Maintenance Proteins and Cdc6 in Mammalian Cells. J Biol Chem 2004;279:9625-33. [Crossref] [PubMed]
- Petropoulou C, Kotantaki P, Karamitros D, et al. Cdt1 and Geminin in cancer: Markers or triggers of malignant transformation? Front Biosci 2008;13:4485-94. [Crossref] [PubMed]
- Borlado LR, Méndez J. CDC6: From DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 2008;29:237-43. [Crossref] [PubMed]
- Karavias D, Maroulis I, Papadaki H, et al. Overexpression of CDT1 Is a Predictor of Poor Survival in Patients with Hepatocellular Carcinoma. J Gastrointest Surg 2016;20:568-79. [Crossref] [PubMed]
- Fan X, Zhou Y, Chen JJ. Role of Cdc6 in re-replication in cells expressing human papillomavirus E7 oncogene. Carcinogenesis 2016;37:799-809. [Crossref] [PubMed]
- Chen S, Chen X, Xie G, et al. Cdc6 contributes to cisplatin-resistance by activation of ATRChk1 pathway in bladder cancer cells. Oncotarget 2016;7:40362-76. [Crossref] [PubMed]
- Bartucci M, Svensson S, Romania P, et al. Therapeutic targeting of Chk1 in NSCLC stem cells during chemotherapy. Cell Death Differ 2012;19:768-78. [Crossref] [PubMed]
- Pintard L, Peter M. Cdc34: Cycling on and off the SCF. Nat Cell Biol 2003;5:856-7. [Crossref] [PubMed]
- Skowyra D, Craig KL, Tyers M, et al. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 1997;91:209-19. [Crossref] [PubMed]
- Butz N, Ruetz S, Natt F, et al. The human ubiquitin-conjugating enzyme Cdc34 controls cellular proliferation through regulation of p27Kip1 protein levels. Exp Cell Res 2005;303:482-93. [Crossref] [PubMed]
- Tanaka K, Kondoh N, Shuda M, et al. Enhanced expression of mRNAs of antisecretory factor-1, gp96, DAD1 and CDC34 in human hepatocellular carcinomas. Biochim Biophys Acta 2001;1536:1-12. [Crossref] [PubMed]
- Han Z, Li Q, Wang Y, et al. Niclosamide induces cell cycle arrest in G1 phase in head and neck squamous cell carcinoma through let-7d/Cdc34 axis. Front Pharmacol 2019;9:1544. [Crossref] [PubMed]
- Franco LC, Morales F, Boffo S, et al. CDK9: A key player in cancer and other diseases. J Cell Biochem 2018;119:1273-84. [Crossref] [PubMed]
- Lolli G, Johnson LN. CAK-Cyclin-Dependent Activating Kinase: A key kinase in cell cycle control and a target for Drugs? Cell Cycle 2005;4:565-70. [Crossref] [PubMed]
- Larochelle S, Merrick KA, Terret ME, et al. Requirements for Cdk7 in the Assembly of Cdk1/Cyclin B and Activation of Cdk2 Revealed by Chemical Genetics in Human Cells. Mol Cell 2007;25:839-50. [Crossref] [PubMed]
- Compe E, Egly JM. Nucleotide Excision Repair and Transcriptional Regulation: TFIIH and beyond. Annu Rev Biochem 2016;85:265-90. [Crossref] [PubMed]
- Coin F, Egly JM. Revisiting the Function of CDK7 in Transcription by Virtue of a Recently Described TFIIH Kinase Inhibitor. Mol Cell 2015;59:513-4. [Crossref] [PubMed]
- Larochelle S, Amat R, Glover-Cutter K, et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 2012;19:1108-15. [Crossref] [PubMed]
- Zhong S, Zhang Y, Yin X, et al. CDK7 inhibitor suppresses tumor progression through blocking the cell cycle at the G2/M phase and inhibiting transcriptional activity in cervical cancer. Onco Targets Ther 2019;12:2137-47. [Crossref] [PubMed]
- Romano G, Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle 2008;7:3664-8. [Crossref] [PubMed]
- Lu H, Xue Y, Yu GK, et al. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. Elife 2015;4:e06535. [Crossref] [PubMed]
- Yin T, Lallena MJ, Kreklau EL, et al. A novel CDK9 inhibitor shows potent antitumor efficacy in preclinical hematologic tumor models. Mol Cancer Ther 2014;13:1442-56. [Crossref] [PubMed]
- Albert TK, Antrecht C, Kremmer E, et al. The establishment of a hyperactive structure allows the tumour suppressor protein p53 to function through P-TEFb during limited CDK9 kinase inhibition. PLoS One 2016;11:e0146648. [Crossref] [PubMed]
- Ko E, Kim Y, Cho EY, et al. Synergistic effect of Bcl-2 and cyclin A2 on adverse recurrence-free survival in stage i non-small cell lung cancer. Ann Surg Oncol 2013;20:1005-12. [Crossref] [PubMed]
- Arsic N, Bendris N, Peter M, et al. A novel function for Cyclin A2: Control of cell invasion via rhoa signaling. J Cell Biol 2012;196:147-62. [Crossref] [PubMed]
- Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248-50. [Crossref] [PubMed]
- Bendris N, Arsic N, Lemmers B, et al. Cyclin A2, Rho GTPases and EMT. Small GTPases 2012;3:225-8. [Crossref] [PubMed]
- Morgan DO. Principles of CDK regulation. Nature 1995;374:131-4. [Crossref] [PubMed]
- Krek W, Nigg EA. Differential phosphorylation of vertebrate p34(cdc2) kinase at the G1/S and G2/M transitions of the cell cycle: Identification of major phosphorylation sites. EMBO J 1991;10:305-16. [Crossref] [PubMed]
- Kedinger V, Meulle A, Zounib O, et al. Sticky siRNAs targeting survivin and cyclin B1 exert an antitumoral effect on melanoma subcutaneous xenografts and lung metastases. BMC Cancer 2013;13:338. [Crossref] [PubMed]
- Kreis NN, Sanhaji M, Krämer A, et al. Restoration of the tumor suppressor p53 by downregulating cyclin B1 in human papillomavirus 16/18-infected cancer cells. Oncogene 2010;29:5591-603. [Crossref] [PubMed]
- Weng L, Du J, Zhou Q, et al. Identification of cyclin B1 and Sec62 as biomarkers for recurrence in patients with HBV-related hepatocellular carcinoma after surgical resection. Mol Cancer 2012;11:39. [Crossref] [PubMed]


