
Bioinformatic analysis of the expression and prognosis of ZNF589 in human breast cancer
Introduction
Breast cancer (BRCA) is the most common type of solid tumour in female cancer patients. The incidence of BRCA varies broadly worldwide and ranges from 27/1,000,002 in Central-East Asia and Africa to 85–94/1,000,002 in Australia, North America and Western Europe (1). The increasing incidence of BRCA has led to the development of an increasing number of choices for diagnosis and therapy. Along with classical biomarkers, such as oestrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), cytokeratin (CK) and the breast cancer 1 (BRCA1) protein, a number of biomarkers of single genes or multigene features have been approved for clinical use for BRCA prognosis. For single genes, hormone receptors, BRCA1/2 and Ki-67 have already been used for pathological reports and molecular subtypes. For multigenes, MammaPrint and Prosigna are newly developed detection methods. Improving the surgical methods and developing more precise radiation therapy and targeted chemotherapy have been well reported as traditional methods, while immunotherapy represents a revolutionary development among the therapeutic methods for BRCA (2). However, almost 12% of BRCA patients eventually develop recurrent or metastatic BRCA, and the prognosis is poor in such patients, and even patients with the same subtype show heterogeneity in prognosis and chemical resistance (3). For example, locally advanced BRCA consists of terminal-stage primary tumours, cancers with extensive lymphatic metastasis and inflammatory BRCA, and it remains a difficult clinical problem in developing countries. Prevention and treatment techniques for locally advanced BRCA have not been developed, and well-coordinated treatment and superior biomarkers remain to be discovered (4). Therefore, the lack of biomarkers with high accuracy, specificity and convenient application is an important issue to be resolved in clinical medicine.
According to the expression data of the TCGA BC cohort, we found that many genes were significantly differentially expressed between normal and BRCA patients, including zinc finger protein 589 (ZNF589), also known as stem cell ZNF1. A study of haematopoietic stem/progenitor cells indicated the possible role of ZNF589 in the differentiation of CD34+ stem cells (5). Cys2-His2 (C2H2)-type zinc finger proteins (ZNFs) are a well-known family of transcription factors and nuclear hormone receptors. Characterized by zinc finger motifs, which are small peptide domains with a secondary structure stabilized by a zinc ion bound to the cysteine and/or histidine residues of the finger, ZNFs play versatile and crucial roles in various biological processes, including development, differentiation, metabolism, autophagy, atrophy, senescence, epithelial-mesenchymal transition (EMT) and apoptosis (6-11). Characterized by an N-terminal Krüppel-associated box domain and a C-terminal array of zinc-finger motifs, Krüppel-associated box (KRAB) domain zinc finger proteins (KZFPs) are the largest subfamily of C2H2-type ZNFs, the largest family of transcriptional regulators in eukaryotes (6,12). KRAB-ZFPs bind to DNA through their tandem zinc finger motifs to act as transcriptional repressors, and gene silencing is conducted by the highly conserved KRAB domain, which recruits histone deacetylase complexes, histone methylases, and heterochromatin proteins (13). Working together with their cofactor tripartite motif protein 28 (TRIM28), KZFPs participate in the transcriptional repression of multiple aspects of development and physiology (14). Even though the remarkable biological features of most KZFPs remain unclear, recent reports have suggested their role in differentiation, cell proliferation, apoptosis and EMT (14-17). According to the first report of ZNF589, it encodes a protein containing C2H2-type zinc fingers and a Krüppel-associated box domain. In the last decade, ZNF589 was sporadically reported in the fields of neurobiology, stem cell biology, immunology and cardiology (13,14,18-21). For oncology, only one report provided a prognostic signature analysis in squamous cell lung carcinoma in The Cancer Genome Atlas (TCGA) database and mentioned stem cell ZNF1 in a 14-gene cluster (22). Thus, ZNF589 may play a role in prognosis in lung cancer. The distinct biological functions of ZNF589 in cancer, especially BRCA, have rarely been reported. Considering the lack of additional biomarkers with high accuracy, specificity and convenience for BRCA diagnosis, therapy and prognosis in clinical medicine as well as the obvious expression differences in the TCGA BC cohort and prognosis in previous bioinformatic preanalyses, this study set out to determine the predictive validity of ZNF589 in BRCA. The study presented here is the first investigation to survey the expression and clinical significance of ZNF589 in BRCA oncogenesis.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-3166).
Methods
First, we used the TCGA, Oncomine and UALCAN to assess the expression of ZNF589. Second, we obtained the survival curve from the Kaplan-Meier (K-M) plotter database and TCGA data collection to assess the prognostic value of ZNF589. To predict the reason for the expressional change in ZNF589, the methylation status of the ZNF589 promoter region and the gene alteration rate (mutation, copy number alteration and so on) of ZNF589 were estimated through UALCAN and cBioPortal, respectively. In terms of related gene and biological process prediction, STRING was used to predict proteins that interact with ZNF589. LinkedOmics was used to identify co-expressing genes with ZNF589. We put these related genes and proteins into their own enrichment modules and used DAVID to obtain the related biological processes and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and this work did not require a statement on obtaining ethical approval because all data were obtained from public databases.
TCGA data collection
TCGA datasets contain sequencing and pathological data on 30 different types of cancers (23). mRNA expression (HiSeqV2) data were obtained from 133 normal samples and 1108 tumour samples. A total of 1108 tumour samples with clinical information were used to assess the prognostic value. The following corresponding clinical characteristics were extracted: age, sex, primary tumour status, regional lymph node status, TNM stage based on the American Joint Committee on Cancer (AJCC), ER status, PR status, HER2 status and histopathologic type. To analyse the overall survival (OS) and relapse-free survival (RFS) of BRCA patients, high- and low-expression groups were differentiated by the median cut-off of ZNF589 expression from 1108 samples and assessed by a K-M survival plot. We used univariate and multivariate Cox proportional hazard regression models to model the prognostic value of ZNF589 for OS and RFS. Age, sex, tumour infiltration, regional lymph node status, TNM stage, ER, PR, HER2 status and histopathologic type were analysed in regression models.
Oncomine
The Oncomine (https://www.oncomine.org/resource/login.html) gene expression microarray database is the largest sequencing analysis database and online data mining tool (24). We used the BRCA vs. normal analysis type and ordered by the underexpression gene rank to analyse target genes. Thresholds were as follows: fold change >1.5, P<0.05 and gene ranking of all.
UALCAN
UALCAN (http://ualcan.path.uab.edu/) is a webtool characterized by a user-friendly, comprehensive and interactive platform. It contains data on 31 types of cancer RNA-seq and clinical information from TCGA (25). We used UALCAN to obtain data from TCGA Dataset-Breast Invasive Cancer to analyse and compare the expression of ZNF589 and methylation of its promoter in tumours and normal tissues. ZNF589 expression and promoter methylation-level analyses based on sample types, cancer stages, patient race, age, BRCA subclasses, tumour histology, patient menopausal status and TP53 mutation status were conducted within this portal. P<0.05 was considered statistically significant.
STRING
STRING (https://string-db.org/) is a database of known and predicted protein-protein interaction (PPI) information covering 5090 kinds of organisms, and it implements Gene Ontology (GO) and KEGG classification systems (26). We constructed a PPI network with the maximum number of interactors to show no more than 20 interactors in the 2nd shell. Of course, we carried out functional enrichment in our node network and highlighted interested item-associated nodes with specific colours.
DAVID
DAVID (https://david.ncifcrf.gov/) is an integrated biological knowledge base aimed at the functional analysis of large gene lists through GO enrichment (27,28). We placed the corresponding genes of the 26 proteins interacting with ZNF589 into the gene list and selected official gene symbols as identifiers. We selected Homo sapiens to limit the annotation and designated it as the background.
cBioPortal
cBioPortal (http://www.cbioportal.org/) is an online web tool for analysing multidimensional tumour genomics data, and we used it to analyse ZNF589 alterations, such as mutations, copy number variations, fusion, amplification and deep deletion (29).
K-M plotter
We used K-M plotter (www.kmplot.com) to assess the prognostic value of ZNF589 on survival in BRCA. Containing 21 types of cancer, K-M plotter includes gene chip and RNA-seq data sources from GEO, TCGA and EGA (30).
LinkedOmics
LinkedOmics (http://www.linkedomics.org/login.php) is an online platform that permits researchers to analyse 32 TCGA cancer-associated and multidimensional datasets (31). We used the invasive breast carcinoma cohort and selected TCGA_BRCA in the HiSeq RNA platform for the search dataset and target dataset, and the Pearson correlation test was used as the statistical method for assessing ZNF589. The LinkFinder module was used to explore ZNF589 co-expression genes. The results are presented graphically in volcano plots, heat maps and scatter plots. The LinkInterpreter module was used with the Gene Set Enrichment Analysis (GSEA) tool to carry out GO biological process enrichment and KEGG pathway enrichment of co-expressed genes of ZNF589. P value was the rank criteria, the minimum number of genes was 3, and 500 simulations were conducted.
Bioinformatics
Bioinformatics (http://www.bioinformatics.com.cn/) is a free online visualization tool for bioinformatics data. We used the Enrichment Dot Bubble module to upload the genes from the LinkedOmics and STRING analysis results. The data format was directed by the online data template.
KEGG
KEGG (https://www.kegg.jp/kegg/) is an online database resource for determining biology functions at the molecular level. We used its pathway module to determine the locations of interesting genes in specific molecular pathways. Target genes were highlighted by the user data mapping function. The KEGG orthologue number of genes was searched in an orthologue table.
GSEA
GSEA is a computational method that checks whether a set of genes are enriched at the top or bottom of a specific ranked list of genes (32,33). We used GSEA 4.0.3 to analyse 20,530 genes in the TCGA genomic matrix grouped by the median ZNF589 expression level. c2.cp.kegg.v7.1.symbols.gmt was selected as the reference gene set, and FDR <0.25 was considered statistically significant.
Statistical analysis
Comparisons of the transcription level of ZNF589 between normal tissue and BRCA groups in the TCGA and Oncomine datasets were conducted through paired/unpaired t-tests. Survival plots from TCGA and K-M plotter were performed by a log-rank test along with the hazard ratio (HR) with 95% confidence interval. To analyse the OS, RFS and post-progression survival of BRCA patients, the samples were split by the median cut-off of ZNF589 expression and assessed by a K-M survival plot along with the HR with 95% confidence interval and log-rank P value. All statistical analyses were performed using SPSS 26.0 software (SPSS, Inc., Chicago, IL, USA), and P<0.05 was considered statistically significant.
Results
Expression level of ZNF589 was downregulated in BRCA patients
To compare the differences in expression between tumour and normal tissue, we analysed the transcriptional level of ZNF589 in unpaired (P<0.0001) (Figure 1A) and paired (P<0.0001) (Figure 1B) samples. We found that ZNF589 was downregulated in BRCA tumour tissue from the TCGA cohort. To test this result in another way, we downloaded the transcriptional data of ZNF589 from the Oncomine database and compared the expression differences between tumour and normal tissues. As expected, we found that the expression of ZNF589 was downregulated in ductal breast carcinoma according to Sorlie breast research (P=0.005) (Figure 1C) and Sorlie breast 2 research (P=0.0019) (Figure 1D), and it was also downregulated in invasive breast carcinoma in Finak breast research (P<0.0001) (Figure 1E). These results indicated that ZNF589 was downregulated in BRCA.
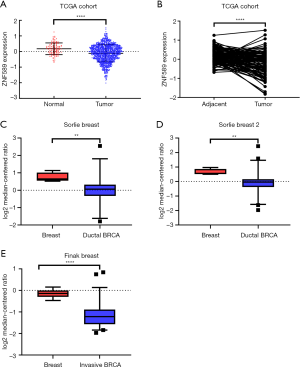
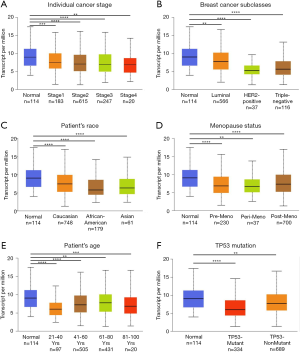
To further explore the expression variance, we used clinical parameters as the classification factors. We found that ZNF589 was downregulated in all cancer stages of BRCA (Figure 2A). For the molecular subclasses, downregulation was found in the luminal, HER2-positive and triple-negative subgroups. In particular, the HER2-positive subtype expressed the lowest level, followed by the triple-negative subtype (luminal vs. HER2-positive, luminal vs. TNBC and HER2-positive vs. TNBC were statistically significant and P value <0.05) (Figure 2B). To explore the correlation between expression and hormone receptor status, we analysed the high- and low-expression rates in different hormone receptor statuses with the chi-square test. We found that the high expression rate of the ER-negative and PR-negative groups was lower as well as the HER2-positive group; in other words, ZNF589 was downregulated in the ER-negative (P<0.001) (Figure S1A), PR-negative (P<0.001) (Figure S1B) and HER2-positive (Figure S1C) groups and ZNF589 expression was correlated with hormone status. To investigate expression differences in different races, we compared three major patients based on race and found that tumour tissues in all races were downregulated; moreover, African Americans and Asians had the lowest levels compared with Caucasians (Caucasian vs. African Americans and Caucasian vs. Asians were statistically significant and P value <0.05) (Figure 2C). We also analysed different menopausal statuses and found statistically significant differences among premenopausal, perimenopausal and postmenopausal patients compared to normal controls (Figure 2D). Moreover, ZNF589 was downregulated in all age groups (Figure 2E), and to further investigate the relationship between expression and patient age, we artificially set the usual age of 55 as the cut-off and defined <55 as the high-risk group and ≥55 as the low-risk group. Interestingly, we found that the high ZNF589 expression rate was upregulated in the low-risk group and that ZNF589 expression was correlated with age status (Figure S1D). We also separated patients into the TP53-mutant group and TP53-nonmutant group. It is universally acknowledged that TP53 mutation correlates to a high risk of oncogenesis. In this case, we found that both groups were downregulated and that the TP53-mutant group was the lowest (Figure 2F). These results indicated that ZNF589 was usually expressed at lower levels in the high-risk subgroup of people.
Downregulation of ZNF589 predicts poor OS in BRCA patients
To investigate the prognostic value of ZNF589, we used the TCGA database to conduct the survival analysis. According to the total OS result, patients with high ZNF589 expression showed a higher OS rate (P=0.047) (Figure 3A). We artificially defined pathological stages I to II as early stages and stages III to IV as terminal stages. Although the OS of the early stage was not obviously changed (P=0.575) (Figure 3B), high ZNF589 led to better OS in terminal stage (P=0.0249) (Figure 3C). As a result, we further explored the survival rate of OS in different hormone statuses to determine the prognosis in specific types. Significant results were observed for PR-negative status (P=0.0002) (Figure 3D) but not ER-negative (P=0.0999) (Figure S2A) or HER2 negative (P=0.6981) (Figure S2B). Considering that OS was different in PR-negative BRCA, we analysed the OS of triple-negative BRCA; however, the number of accessible cases was too low to analyse. For triple-negative BRCA, it overlapped with basal-like BRCA. We used K-M plotter to analyse the association between ZNF589 expression and the clinical outcome in OS of basal-like BRCA. Surprisingly, the result was statistically significant (Figure S2C).
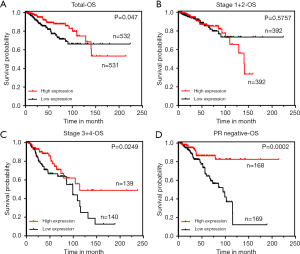
Furthermore, univariate and multivariate Cox regression models were used to assess the prognostic value of ZNF589 (Table 1). What stood out in the table was that ZNF589 expression (low vs. high) was one of independent prognostic factors (HR =0.56, P=0.021), which included age (≤55 vs. >55 years) (HR =1.99, P=0.008), lymph node metastasis (N0 vs. N1–3) (P<0.001), distant metastasis (M0 vs. M1) (P<0.001) and AJCC TNM stage (I–II vs. III–IV) (HR =2.14, P=0.003). These results showed that ZNF589 could act as an independent prognostic biomarker in BRCA.
Table 1
| Clinicopathologic parameters | OS | ||||||
|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (≤55 vs. >55 years) | 1.67 | 1.17–2.38 | 0.005** | 1.99 | 1.20–3.30 | 0.008** | |
| Histological (IDC vs. ILC) | 1.56 | 0.91–2.69 | 0.109 | ||||
| ER status (positive vs. negative) | 0.72 | 0.49–1.06 | 0.097 | ||||
| PR status (positive vs. negative) | 0.71 | 0.49–1.02 | 0.061 | ||||
| HER2 status (positive vs. negative) | 1.05 | 0.57–1.94 | 0.869 | ||||
| Tumour infiltration (T1–T2 vs. T3–T4) | 1.23 | 0.81–1.85 | 0.336 | ||||
| Lymph node metastasis (N0 vs. N1–3) | 2.11 | 1.44–3.10 | 0.000*** | ||||
| Distant metastasis (M0 vs. M1) | 3.82 | 2.13–6.84 | 0.000*** | ||||
| TNM stage (I–II vs. III–IV) | 2.01 | 1.40–2.90 | 0.000*** | 2.14 | 1.31–3.51 | 0.003** | |
| ZNF589 expression (low vs. high)a | 0.69 | 0.49–0.98 | 0.038* | 0.56 | 0.34–0.92 | 0.021* | |
a, using median H-score values as cutoff. *, P<0.05; **, P<0.01; ***, P<0.001. OS, overall survival; BRCA, breast cancer; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; ER, oestrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Downregulation of ZNF589 predicts poor RFS in BRCA patients
To further explore the prognostic value of ZNF589 in BRCA, we analysed the association between ZNF589 expression and clinical outcomes in RFS (P=0.0043) (Figure 4A). The results showed that high expression of ZNF589 predicted better RFS in BRCA patients. Similarly, we artificially defined the early stage and the terminal stage as OS. Although the RFS of the early stage was not obviously distinguished (P=0.1580) (Figure 4B), high ZNF589 led to better RFS in the terminal stage (P=0.0090) (Figure 4C). In these cases, we explored RFS in different histological subtypes and found that high ZNF589 expression significantly predicted better prognosis in invasive ductal carcinoma (IDC) (P=0.0091) (Figure S2D). While the RFS of the IDC early stage was not obviously significant (P=0.5214) (Figure S2E), high ZNF589 corresponded to better RFS in terminal stage (P=0.0045) (Figure S2F). Hormone status was also considered in the RFS analysis. The single most striking observation to emerge from the data comparison was that better prognosis was found in ER-negative (P=0.0129) (Figure 4D), PR-negative (P=0.0130) (Figure 4E) and HER2-negative BRCA (P=0.00002) (Figure S2G). The triple-negative molecular subtype was assessed, and the same trend was statistically significant (P=0.0052) (Figure 4F). These results indicated that ZNF589 is a potential prognostic factor, especially in advanced stages, and has IDC subtype specificity in RFS.
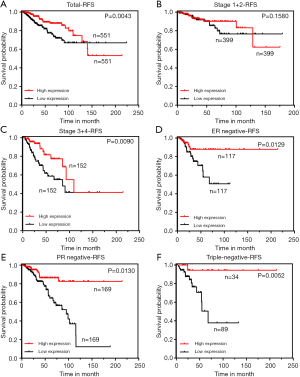
For RFS, univariate and multivariate Cox regression models were again used to assess the prognostic value of ZNF589 (Table 2) and revealed that ZNF589 expression (low vs. high) was one of the independent prognostic factors (HR =0.43, P=0.017), which included PR status (positive vs. negative) (P=0.015), tumour infiltration (T1–T2 vs. T3–T4) (P=0.047), lymph node metastasis (N0 vs. N1–3) (P<0.001), distant metastasis (M0 vs. M1) (HR =5.06, P=0.003) and AJCC TNM stage (I–II vs. III–IV) (P<0.001).
Table 2
| Clinicopathologic parameters | RFS | ||||||
|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (≤55 vs. >55 years) | 1.10 | 0.71–1.71 | 0.680 | ||||
| Histological (IDC vs. ILC) | 1.38 | 0.74–2.57 | 0.313 | ||||
| ER status (positive vs. negative) | 0.63 | 0.39–1.02 | 0.059 | ||||
| PR status (positive vs. negative) | 0.57 | 0.36–0.90 | 0.015* | ||||
| HER2 status (positive vs. negative) | 0.69 | 0.28–1.73 | 0.429 | ||||
| Tumour infiltration (T1–T2 vs. T3–T4) | 1.67 | 1.01–2.78 | 0.047* | ||||
| Lymph node metastasis (N0 vs. N1–3) | 2.56 | 1.55–4.23 | 0.000*** | ||||
| Distant metastasis (M0 vs. M1) | 5.96 | 2.56–13.86 | 0.000*** | 5.06 | 1.76–14.50 | 0.003** | |
| TNM stage (I–II vs. III–IV) | 2.83 | 1.79–4.49 | 0.000*** | ||||
| ZNF589 expression (low vs. high)a | 0.50 | 0.31–0.79 | 0.003** | 0.43 | 0.22–0.86 | 0.017* | |
a, using median H-score values as cutoff. *, P<0.05; **, P<0.01; ***, P<0.001. RFS, relapse-free survival; BRCA, breast cancer; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; ER, oestrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
ZNF589 co-expressive genes for probable mechanism in BRCA
To investigate the possible mechanism of oncogenic suppression, we explored ZNF589 co-expression genes through the LinkedOmics database and found that there were 6,904 positively correlated genes and 4,622 negatively correlated genes (FDR <0.05). We then obtained a heatmap of the top 50 positively and negatively correlated genes from a volcano plot of all ZNF589-associated genes (Figure 5A,B,C). The matched module of LinkedOmics was used to conduct a GSEA to determine the KEGG pathways of associated genes (Figure 5D). To further confirm the previous results, we carried out a GSEA of ZNF589 with KEGG gene sets in BRCA based on the TCGA cohort. There were 1,215 samples and 20,530 genes in the analysis. We divided the 1,215 samples into a High Group and Low Group by the median cut-off value of ZNF589. No significantly enriched gene set was found in the High (H) phenotype, but 8 gene sets were significantly enriched in the Low (L) phenotype at a nominal P value <1%, and 19 gene sets were significantly enriched at a nominal P value <5%. Comparing the results of the LinkedOmics matched GSEA and TCGA cohort GSEA enrichment analysis, we found that there were many KEGG pathways in common and related to tumorigenesis. Because the cell cycle is a conventional research aspect in BRCA, we focused on the cell cycle pathway for the convenience of subsequent experimental research. Remarkably, the cell cycle gene set [|Normalized Enrichment Score (NES)| >1, NOM P value <0.05, FDR q value <0.25] was negatively correlated with ZNF589 expression (Figure 5E). There were 50 core enriched genes in the cell cycle gene set (Table S1). For the cell cycle from the KEGG gene set, the top three leading-edge genes were cyclin E1 (CCNE1), polo-like kinase 1 (PLK1), and cyclin B2 (CCNB2). CCNE1 is a member of the cyclin E family and positively related to the G1/S phase transition (34). It has already been demonstrated that deregulated cyclin E induces chromosome instability and drives cells towards more advanced tumour stages (35). PLK1 is a key mediator of the transition of the G2-M phase of the cell cycle, and its inhibition induces mitotic arrest of BRCA (36,37). CCNB2 was first reported to be associated with BRCA prognosis in 2013 and is a member of the cyclin family controlling the G2/M phase transition (38). To provide a better understanding of the location and function of leading-edge genes in the gene set of the cell cycle pathway, we placed the leading-edge genes into the KEGG pathway mapping module (Figure 5F). The results showed that ZNF589 could play an important role during breast tumorigenesis, probably through cell cycle regulation.
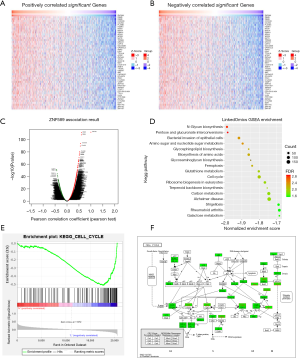
Genome-wide proteins associated with ZNF589 and probable mechanism in BRCA
To explore the possible role of ZNF589 as a tumour suppressor at the protein level, we used the STRING database to analyse the PPI network. As shown in the network, the predicted functional partners were TRIM28, clavesin-1 (CLVS1), ZNF618, NEDD4 binding protein 2 like 1 (N4BP2L1) and transmembrane protein 92 (TMEM92). Considering that only 4 functional partners were predicted, we added the second shell of nodes in the analysis and obtained more indirectly interactive proteins: SUMO-conjugating enzyme UBC9 (UBE2I), E3 ubiquitin-protein ligase Mdm2 (MDM2), cellular tumour antigen p53 (TP53), paired amphipathic helix protein Sin3a (SIN3A), serine-protein kinase ATM (ATM), mediator of DNA damage checkpoint protein 1 (MDC1), TP53-binding protein 1 (TP53BP1), Histone H2AX (H2AFX), histone deacetylase 1 (HDAC1), small ubiquitin-related modifier 1 (SUMO1), cell division cycle 5-like protein (CDC5L), ZNF350, ZNF274, ZNF with KRAB and SCAN domains 7 (ZKSCAN7), etc. (Figure 6A). To determine which BP and KEGG pathways were enriched in these proteins, we performed a web-matched enrichment analysis to obtain a summary of KEGG pathways. We found that these proteins were also related to pathways involved in the cell cycle, and we highlighted these nodes with different colours for each item (Figure 6A). To further confirm this result, we input these interactive protein-related genes into the DAVID online annotation tool to conduct GO enrichment analyses, and important enriched pathways were identified (Figure 6B). Surprisingly, we obtained the cell cycle pathway again, and the leading-edge genes HDAC1, TP53, MDM2 and ATM were placed into the KEGG pathway through the DAVID mapping tool (Figure 6C). These results strongly indicated that ZNF589 probably regulates the cell cycle through a protein-interactive mechanism.
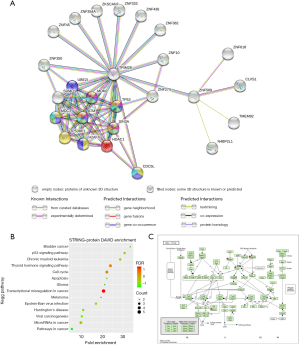
Downregulation of ZNF589 transcription associated with hypermethylation of its promoter region but not genetic alteration in BRCA
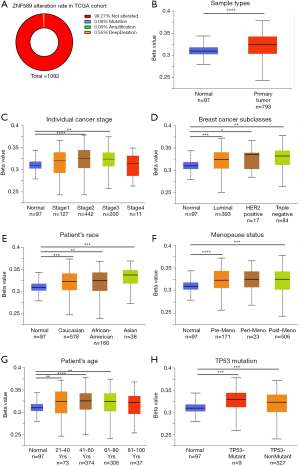
To determine the reason for ZNF589 downregulation, we investigated the alteration status through cBioPortal (Figure 7A) and hypermethylation of the ZNF589 promoter area through UALCAN. We found that mutations, amplifications and deep deletions of ZNF589 were rare. Statistically, the alteration rate was extremely low. The changes in the relationship between the expression of ZNF589 and its promoter methylation level in the TCGA samples were compared. A significant outcome was obtained: the expression of the primary tumour was downregulated while the promoter methylation level was upregulated (Figure 7B). As the pathological stage advanced, the expression of ZNF589 was downregulated compared to that in normal tissues and the methylation level of ZNF589 was upregulated (Figure 7C). Specifically, the methylation level of all tumour subclasses was upregulated (Figure 7D). Regarding racial differences, we found the same trend: when the expression of the primary tumour was downregulated, the promoter methylation level was upregulated. We also observed that Asian patients had the highest methylation (Caucasian vs. Asian and African-American vs. Asian were statistically significant and P value <0.05) (Figure 7E). This finding suggested that there might be a racial component to the promoter methylation level of ZNF589. In addition to racial elements and tumour subclasses, we also considered menopausal status (Figure 7F), patient age (Figure 7G) and TP53 mutation (Figure 7H). We observed that the promoter methylation level was upregulated while the expression of the primary tumour was downregulated. We speculated that the demethylation of the ZNF589 promoter area could be a therapeutic target for BRCA patients with low ZNF589 expression.
Discussion
ZNF589 is a member of the ZNF family, and its encoded product contains C2H2-type ZNFs and KRAB (5). Its role in tumorigenesis is seldom reported. Herein, we assessed the latent role of ZNF589 in breast carcinogenesis. Our study found for the first time that epigenetic silencing of ZNF589 was associated with poor OS and RFS in BRCA. The Cox regression results suggested that ZNF589 could act as an independent prognostic biomarker for BRCA. Further analysis showed that ZNF589 expression was negatively correlated with cell cycle progression at the transcriptional and protein-interactive levels.
Analysis of the transcriptome from different databases for all kinds of parameters confirmed that ZNF589 was downregulated in tumour tissues, which suggested that ZNF589 might be a tumour suppressor (39). The subgroup analysis divided by cancer stage showed that there was a gradual decline in ZNF589 expression, which might be negatively correlated with stage progression or malignancy to some degree. According to the subgroup divided by hormone status, the HER2-positive subtype expressed the lowest level, followed by the triple-negative subtype. Interestingly, HER2 positivity and TNBC are malignant subtypes in clinical practice. Therefore, we assumed that ZNF589 expression was negatively correlated with BRCA malignancy. Furthermore, the correlation analysis indicated that the expression of ZNF589 was downregulated in ER-negative, PR-negative and HER2-positive cells and negatively related to BRCA malignancy. In terms of race, the race-specific expression module indicated that the role of ZNF589 might be more significant in specific human races. The menopause results indicated that hormone level changes did not affect the ZNF589 low-expression state.
Low expression of ZNF589 was significantly related to poor OS and RFS (40). The survival trend in OS showed that ZNF589 might be a potential tumorigenic suppressor in BRCA. On the other hand, the survival trend in RFS implied that ZNF589 may play a key role in the suppression of local tumour recurrence. Thus, ZNF589 may play a role in cell proliferation, apoptosis, senescence, pyroptosis and the cell cycle (41-45). Interestingly, OS was statistically significant in basal-like BRCA, while RFS was statistically significant in triple-negative and IDC subtype BRCA. These results hinted that ZNF589 could also play a role in the overall prognosis and local recurrence of specific subtypes of BRCA. However, how ZNF589 acts on tumorigenesis and recurrence remains unclear.
To explore the possible mechanism of ZNF589 in oncogenic repression, we conducted a LinkedOmics matched GSEA and TCGA cohort GSEA enrichment analysis, and we found there were many KEGG pathways in common and related to tumorigenesis, including the proteasome (46,47), oxidative phosphorylation (48,49), vibrio cholerae infection (50,51), Parkinson disease (52,53), protein export (54,55), pathogenic Escherichia coli infection (56,57), N-Glycan biosynthesis (58,59), glycosphingolipid biosynthesis (60,61), glutathione metabolism (62,63), cell cycle (64,65), Alzheimer disease (66,67) and galactose metabolism pathways (68,69). Transcriptionally, we obtained the cell cycle pathway as one of the enriched results, and the leading-edge subset of the cell cycle was upregulated in the ZNF589 low-expression group. In other words, low expression of ZNF589 was accompanied by high expression of leading-edge genes that promoted the cell cycle process. CCNE1 is mediated by the ZNF family to regulate the G1 restriction point (70). Therefore, we speculated that ZNF589 could possibly transcriptionally repress the expression of CCNE1 and other GSEA leading-edge genes associated with the cell cycle to block the tumour cell cycle in BRCA. At the protein-interactive level, we know that ZNF589 was co-expressed with TRIM28 from curated databases, text mining and experimentally determined methods. With the STRING matched online enrichment-analysis tool and DAVID online annotation tool, microRNAs in the cancer (71,72), cell cycle and p53 signalling pathways were enriched, and they are all well-known cancer-related pathways. We obtained the cell cycle KEGG pathway, and biological processes were enriched. As a result, ZNF589 may also regulate the cell cycle process through TRIM28 via a protein-interactive method. Thus, we hypothesize that TRIM28 plays a key role in BRCA repression of ZNF589. First, ZNF589 could combine with TRIM28 to transcriptionally regulate downstream genes to arrest the cell cycle. For example, TRIM28 acts as a nuclear corepressor with KZFPs to mediate gene silencing by recruiting chromodomain helicase DNA binding protein 3 (CHD3), which is a subunit of the nucleosome remodelling and deacetylation (NuRD) complex, and SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) specifically methylates histone H3 at ‘Lys-9’ (H3K9me) to the promoter regions of KRAB target genes (14). TRIM28 enhances transcriptional repression by coordinating the increase in H3K9me, the decrease in histone H3 ‘Lys-9’ and ‘Lys-14’ acetylation (H3K9ac and H3K14ac, respectively) and the disposition of heterochromatin protein 1 (HP1) proteins (73,74). Therefore, KZFPs work with TRIM28 to conduct transcriptional repression. Coincidently, ZNF589 was first found to encode a protein containing C2H2-type zinc finger motifs and a Kruppel-associated box (KRAB) domain in a study of haematopoietic stem/progenitor cells (5). Therefore, we assumed that ZNF589 combined with TRIM28 to transcriptionally repress downstream genes. Interestingly, not all STRING-predicted proteins promoted the cell cycle, such as TP53 and SIN3A. Although the complex suppression of these antitumour genes by ZNF589-TRIM28 may contradict the tumour repressing role of ZNF589, the ZNF589-TRIM28 complex might also transcriptionally promote the expression of downstream proteins to repress the cell cycle in an unknown way. Alternatively, ZNF589 probably cooperates with TRIM28 to induce PPI with downstream proteins to repress the cell cycle. TRIM28 itself is an oncogenic gene and promotes proliferation and metastatic progression in BRCA (75). Thus, ZNF589 could possibly regulate the transcription of TRIM28 to exert a tumour-suppressive role.
After exploring the possible oncogenic repression mechanism, we found that ZNF589 was a possible tumour suppressor and a promising biomarker for BRCA. To clarify the reason for the downregulation of ZNF589, we took genetic and epigenetic levels into consideration. Although gene alterations of ZNF589 were rare in BRCA in cBioPortal, we found that the downregulation of ZNF589 was accompanied by high methylation in the ZNF589 promoter area, which was consistent with the comparison of the UALCAN expression analysis and methylation analysis of ZNF589 in BRCA. Thus, downregulation may not have been caused by gene alteration in the DNA sequence of ZNF589 but by methylation of the ZNF589 transcriptional promoter sequence.
Limitations
We used several online analysis tools to obtain the statistics about ZNF589 and reconfirmed these data to ensure the reliability. However, the lack of experimental data introduced uncertainty in our results. Clinical patient tissue could be used to test the mRNA and protein expressional variance of ZNF589 in tumour and normal tissues.
In this study, we systematically analysed the expression and prognostic value of ZNF589 in BRCA. Our analysis predicted that ZNF589 was downregulated and that its promoter region was highly methylated in BRCA patients, especially in terminal patients. Moreover, low expression of ZNF589 was accompanied by poor OS and RFS. Therefore, ZNF589 might be a latent prognostic biomarker in terminal BRCA. Regarding the repressive mechanism of tumorigenesis, ZNF589 may suppress cancer through many important cancer-associated pathways, such as the cell cycle. In conclusion, ZNF589 may be a latent terminal-stage prognostic biomarker and potential treatment target for BRCA patients.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-3166
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3166). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and this article is not required a statement that ethical approval was obtained because all data was obtained from public database.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sancho-Garnier H, Colonna M. Breast cancer epidemiology. Presse Med 2019;48:1076-84. [Crossref] [PubMed]
- Emens LA. Breast cancer immunotherapy: facts and hopes. Clin Cancer Res 2018;24:511-20. [Crossref] [PubMed]
- Peart O. Metastatic breast cancer. Radiol Technol 2017;88:519M-39M. [PubMed]
- Tryfonidis K, Senkus E, Cardoso MJ, et al. Management of locally advanced breast cancer-perspectives and future directions. Nat Rev Clin Oncol 2015;12:147-62. [Crossref] [PubMed]
- Liu C, Levenstein M, Chen J, et al. SZF1: a novel KRAB-zinc finger gene expressed in CD34+ stem/progenitor cells. Exp Hematol 1999;27:313-25. [Crossref] [PubMed]
- Ecco G, Imbeault M, Trono D. KRAB zinc finger proteins. Development 2017;144:2719-29. [Crossref] [PubMed]
- Lynch SA, McLeod MA, Orsech HC, et al. Zinc finger protein 593 is upregulated during skeletal muscle atrophy and modulates muscle cell differentiation. Exp Cell Res 2019;383:111563. [Crossref] [PubMed]
- Xiang Q, Zhou D, He X, et al. The zinc finger protein GATA4 induces mesenchymal-to-epithelial transition and cellular senescence through the nuclear factor-κB pathway in hepatocellular carcinoma. J Gastroenterol Hepatol 2019;34:2196-205. [Crossref] [PubMed]
- Liang Y, Li Q, Chen K, et al. Zinc finger protein 307 functions as a tumor-suppressor and inhibits cell proliferation by inducing apoptosis in hepatocellular carcinoma. Oncol Rep 2017;38:2229-36. [Crossref] [PubMed]
- Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci 2016;23:53. [Crossref] [PubMed]
- Gamsjaeger R, Liew CK, Loughlin FE, et al. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 2007;32:63-70. [Crossref] [PubMed]
- Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet 2009;5:e1000325. [Crossref] [PubMed]
- Peng H, Zheng L, Lee WH, et al. A common DNA-binding site for SZF1 and the BRCA1-associated zinc finger protein, ZBRK1. Cancer Res 2002;62:3773-81. [PubMed]
- Oleksiewicz U, Gładych M, Raman AT, et al. TRIM28 and Interacting KRAB-ZNFs Control Self-Renewal of Human Pluripotent Stem Cells through Epigenetic Repression of Pro-differentiation Genes. Stem Cell Reports 2017;9:2065-80. [Crossref] [PubMed]
- Huang C, Wu S, Li W, et al. Zinc-finger protein p52-ZER6 accelerates colorectal cancer cell proliferation and tumour progression through promoting p53 ubiquitination. EBioMedicine 2019;48:248-63. [Crossref] [PubMed]
- Tian C, Xing G, Xie P, et al. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 2009;11:580-91. [Crossref] [PubMed]
- Pei L, He X, Li S, et al. KRAB zinc-finger protein 382 regulates epithelial-mesenchymal transition and functions as a tumor suppressor, but is silenced by CpG methylation in gastric cancer. Int J Oncol 2018;53:961-72. [Crossref] [PubMed]
- Lee CI, Chou AK, Lin CC, et al. Immune and inflammatory gene signature in rat cerebrum in subarachnoid hemorrhage with microarray analysis. Mol Med Rep 2012;5:118-25. [PubMed]
- Agha Z, Iqbal Z, Azam M, et al. Exome sequencing identifies three novel candidate genes implicated in intellectual disability. PLoS One 2014;9:e112687. [Crossref] [PubMed]
- Mo XB, Lei SF, Zhang YH, et al. Examination of the associations between m6A-associated single-nucleotide polymorphisms and blood pressure. Hypertens Res 2019;42:1582-9. [Crossref] [PubMed]
- Venturini L, Stadler M, Manukjan G, et al. The stem cell zinc finger 1 (SZF1)/ZNF589 protein has a human-specific evolutionary nucleotide DNA change and acts as a regulator of cell viability in the hematopoietic system. Exp Hematol 2016;44:257-68. [Crossref] [PubMed]
- Li J, Wang J, Chen Y, et al. A prognostic 4-gene expression signature for squamous cell lung carcinoma. J Cell Physiol 2017;232:3702-13. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Luo Q, Li X, Li J, et al. MiR-15a is underexpressed and inhibits the cell cycle by targeting CCNE1 in breast cancer. Int J Oncol 2013;43:1212-8. [Crossref] [PubMed]
- Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature 1999;401:297-300. [Crossref] [PubMed]
- Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol 2004;5:429-40. [Crossref] [PubMed]
- Maire V, Némati F, Richardson M, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res 2013;73:813-23. [Crossref] [PubMed]
- Shubbar E, Kovács A, Hajizadeh S, et al. Elevated cyclin B2 expression in invasive breast carcinoma is associated with unfavorable clinical outcome. BMC Cancer 2013;13:1. [Crossref] [PubMed]
- Song Y, Zhao Y, Ding X, et al. microRNA-532 suppresses the PI3K/Akt signaling pathway to inhibit colorectal cancer progression by directly targeting IGF-1R. Am J Cancer Res 2018;8:435-49. [PubMed]
- Bruun J, Sveen A, Barros R, et al. Prognostic, predictive, and pharmacogenomic assessments of CDX2 refine stratification of colorectal cancer. Mol Oncol 2018;12:1639-55. [Crossref] [PubMed]
- Xu H, Jiao Y, Yi M, et al. EYA2 correlates with clinico-pathological features of breast cancer, promotes tumor proliferation, and predicts poor survival. Front Oncol 2019;9:26. [Crossref] [PubMed]
- Pan L, Li Y, Zhang HY, et al. DHX15 is associated with poor prognosis in acute myeloid leukemia (AML) and regulates cell apoptosis via the NF-kB signaling pathway. Oncotarget 2017;8:89643-54. [Crossref] [PubMed]
- Miettinen TP, Peltier J, Härtlova A, et al. Thermal proteome profiling of breast cancer cells reveals proteasomal activation by CDK4/6 inhibitor palbociclib. EMBO J 2018;37:e98359. [Crossref] [PubMed]
- Chen C, Wang B, Sun J, et al. DAC can restore expression of NALP1 to suppress tumor growth in colon cancer. Cell Death Dis 2015;6:e1602. [Crossref] [PubMed]
- Zhao L, Jiang L, He L, et al. Identification of a novel cell cycle-related gene signature predicting survival in patients with gastric cancer. J Cell Physiol 2019;234:6350-60. [Crossref] [PubMed]
- Guo X, Wang X, Wang Z, et al. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat Cell Biol 2016;18:202-12. [Crossref] [PubMed]
- Walerych D, Lisek K, Sommaggio R, et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol 2016;18:897-909. [Crossref] [PubMed]
- Lee KM, Giltnane JM, Balko JM, et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab 2017;26:633-47.e7. [Crossref] [PubMed]
- LeBleu VS, O'Connell JT, Gonzalez Herrera KN, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 2014;16:992-1003, 1-15.
- Ray T, Kar D, Pal A, et al. Molecular targeting of breast and colon cancer cells by PAR1 mediated apoptosis through a novel pro-apoptotic peptide. Apoptosis 2018;23:679-94. [Crossref] [PubMed]
- Ray T, Pal A. PAR-1 mediated apoptosis of breast cancer cells by V. cholerae hemagglutinin protease. Apoptosis 2016;21:609-20. [Crossref] [PubMed]
- Lin PY, Chang SN, Hsiao TH, et al. Association Between Parkinson Disease and Risk of Cancer in Taiwan. JAMA Oncol 2015;1:633-40. [Crossref] [PubMed]
- Liu J, Zhang C, Zhao Y, et al. Parkin targets HIF-1α for ubiquitination and degradation to inhibit breast tumor progression. Nat Commun 2017;8:1823. [Crossref] [PubMed]
- Rezaeian AH, Li CF, Wu CY, et al. A hypoxia-responsive TRAF6-ATM-H2AX signalling axis promotes HIF1α activation, tumorigenesis and metastasis. Nat Cell Biol 2017;19:38-51. [Crossref] [PubMed]
- Li L, Hanahan D. Hijacking the neuronal NMDAR signaling circuit to promote tumor growth and invasion. Cell 2013;153:86-100. [Crossref] [PubMed]
- Onda M, Wang QC, Guo HF, et al. In vitro and in vivo cytotoxic activities of recombinant immunotoxin 8H9(Fv)-PE38 against breast cancer, osteosarcoma, and neuroblastoma. Cancer Res 2004;64:1419-24. [Crossref] [PubMed]
- Friedman PN, McAndrew SJ, Gawlak SL, et al. BR96 sFv-PE40, a potent single-chain immunotoxin that selectively kills carcinoma cells. Cancer Res 1993;53:334-9. [PubMed]
- Kyselova Z, Mechref Y, Kang P, et al. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clin Chem 2008;54:1166-75. [Crossref] [PubMed]
- Terkelsen T, Haakensen VD, Saldova R, et al. N-glycan signatures identified in tumor interstitial fluid and serum of breast cancer patients: association with tumor biology and clinical outcome. Mol Oncol 2018;12:972-90. [Crossref] [PubMed]
- Battula VL, Shi Y, Evans KW, et al. Ganglioside GD2 identifies breast cancer stem cells and promotes tumorigenesis. J Clin Invest 2012;122:2066-78. [Crossref] [PubMed]
- Chuang PK, Hsiao M, Hsu TL, et al. Signaling pathway of globo-series glycosphingolipids and β1,3-galactosyltransferase V (β3GalT5) in breast cancer. Proc Natl Acad Sci U S A 2019;116:3518-23. [Crossref] [PubMed]
- Luo M, Shang L, Brooks MD, et al. Targeting breast cancer stem cell state equilibrium through modulation of redox signaling. Cell Metab 2018;28:69-86.e6. [Crossref] [PubMed]
- Lien EC, Lyssiotis CA, Juvekar A, et al. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat Cell Biol 2016;18:572-8. [Crossref] [PubMed]
- Qin J, Zhou Z, Chen W, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun 2015;6:8471. [Crossref] [PubMed]
- O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol 2016;13:417-30. [Crossref] [PubMed]
- Pathak GA, Zhou Z, Silzer TK, et al. Two-stage Bayesian GWAS of 9576 individuals identifies SNP regions that are targeted by miRNAs inversely expressed in Alzheimer's and cancer. Alzheimers Dement 2020;16:162-77. [Crossref] [PubMed]
- Feng YA, Cho K, Lindstrom S, et al. Investigating the genetic relationship between Alzheimer's disease and cancer using GWAS summary statistics. Hum Genet 2017;136:1341-51. [Crossref] [PubMed]
- Cao Q, Chen X, Wu X, et al. Inhibition of UGT8 suppresses basal-like breast cancer progression by attenuating sulfatide-αVβ5 axis. J Exp Med 2018;215:1679-92. [Crossref] [PubMed]
- Liao JH, Chien CT, Wu HY, et al. A multivalent marine lectin from crenomytilus grayanus possesses anti-cancer activity through recognizing globotriose Gb3. J Am Chem Soc 2016;138:4787-95. [Crossref] [PubMed]
- Pozner A, Terooatea TW, Buck-Koehntop BA. Cell-specific Kaiso (ZBTB33) Regulation of Cell Cycle through Cyclin D1 and Cyclin E1. J Biol Chem 2016;291:24538-50. [Crossref] [PubMed]
- Cai WL, Huang WD, Li B, et al. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol Cancer 2018;17:9. [Crossref] [PubMed]
- Yan W, Wu X, Zhou W, et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol 2018;20:597-609. [Crossref] [PubMed]
- Schultz DC, Friedman JR, Rauscher FJ 3rd. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 2001;15:428-43. [Crossref] [PubMed]
- Ivanov AV, Peng H, Yurchenko V, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 2007;28:823-37. [Crossref] [PubMed]
- Addison JB, Koontz C, Fugett JH, et al. KAP1 promotes proliferation and metastatic progression of breast cancer cells. Cancer Res 2015;75:344-55. [Crossref] [PubMed]


