
Prognostic and diagnostic value of SOX9 in cirrhotic and noncirrhotic hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) has been recognized as a highly malignant tumor that causes a heavy disease burden in humans (1). Many clinical studies have been conducted to explore the potential risk factors of tumor recurrence and overall survival (OS) (2). Liver cirrhosis has long been identified as one of the independent risk factors of tumor recurrence and OS (3). Depending on the presence of cirrhosis, HCC can be classified into cirrhotic HCC (CHCC) and noncirrhotic HCC (NCHCC). Previous studies found that CHCC and NCHCC were distinct entities with respect to sex, age, etiology, tumor related characteristics (size of nodule and extrahepatic metastasis, laboratory examination results), and prognosis (4-6). Furthermore, diagnostic methods and prognostic factors might be different between CHCC and NCHCC, but few studies have been designed to clarify this issue (7,8).
Sex-Determining Region Y-Box 9 (SOX9) is a transcription factor closely related to sex determination. Previous studies have shown that abnormal expression of SOX9 is closely related to the occurrence and malignant progression of tumors. In the prostate, it has been reported that high expression of SOX9 promotes tumor occurrence (9). In breast cancer, SOX9 regulates the expression of LRP6 and TCF4 to activate Wnt signaling and promote tumor proliferation, invasion and metastasis (10). In HCC, SOX9 is also involved in the regulation of tumorigenicity, and its expression is prognosis-related (11). One study (12) reported that SOX9 functions as a marker of cell stemness in cancer stem cell (CSC) in HCC, and forced SOX9 expression restores the self-renewal capacity of non-CSCs. These studies strongly support the important roles of SOX9 in tumor occurrence and progression. However, whether the expression and clinical significance of SOX9 is different between CHCC and NCHCC is still not clear.
Therefore, this study was designed to explore the prognostic and diagnostic value of SOX9 in both CHCC and NCHCC. To our knowledge, this study is the first to explore the value of SOX9 in CHCC and NCHCC using RNA sequencing, quantitative real-time polymerase chain reaction (qRT-PCR) and immunohistochemistry (IHC). The findings of this study will be helpful to clarify similarities and differences between CHCC and NCHCC. We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) checklist (available at https://dx.doi.org/10.21037/tcr-20-3385).
Methods
Patient selection
RNA sequencing data were downloaded from The Cancer Genome Atlas (TCGA). Clinicopathological data and expression values of SOX9 were extracted for further analysis. Extracted data include age, sex, cirrhotic status, Child-Pugh classification, family cancer history, serum α-fetoprotein (AFP) level, pathologic stage, T stage, N stage, M stage, recurrence, and vessels in the tumor. In TCGA, 374 HCC tissues and 50 adjacent normal tissues exhibited SOX9 expression. After patients with missing data on cirrhotic status were excluded, 79 CHCC and 133 NCHCC patients were obtained for analysis. Clinical and pathological traits of these patients are summarized in Table S1. Additionally, in our study, 99 pairs of fresh HCC tissues and adjacent normal tissues were obtained from patients who underwent liver resection in the Second Affiliated Hospital of Chongqing Medical University. All diagnoses were histopathologically confirmed. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics committee of the Second Affiliated Hospital of Chongqing Medical University (NO.2020-329) and informed consent was taken from all the patients.
qRT-PCR
Total RNA was extracted from liver tissue using the classic TRIzol method. Afterward, the concentration and purity of RNA were measured. After confirming the quality of total RNA, the RNA underwent reverse transcription using the iScriptTM gDNA Clear cDNA Synthesis Kit (Bio-Rad, United States). Furthermore, with the cDNA samples, qRT-PCR was conducted for quantification using the SsoFastTM EvaGreen® Supermix Kit (Bio-Rad, United States). Expression of SOX9 was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Relative RNA expression was obtained using the 2−∆∆Ct method.
IHC
Formalin-fixed, paraffin-embedded tissues were used for IHC analysis to assess SOX9 expression. The SOX9 antibody (Abcam, United Kingdom) was diluted to desired concentrations and incubated at room temperature. Detection was then accomplished using the 3,3’-diaminobenzidine (DAB) method. IHC results were scored based on color depth and percentage of positive cells. Based on the color depth in the tissue array, scores were 0, 1, 2, and 3, representing negative, canary yellow, pale brown, and dark brown, respectively. Moreover, with respect to the percentage of positive cells, scores were 1, 2, 3, and 4, representing 0–25%, 26–50%, 51–75%, and >75%, respectively. For each tissue, the score was the product of the color depth score and the cell percentage score.
Statistical analysis
SPSS software (version 22.0; IBM, United States) and Stata (version 16.0; stata, United States) were used for statistical analysis. GraphPad prism (version 7.0a; GraphPad Software, United States) was used for graphical display. Student’s t-test or the Mann-Whitney test was used for comparison of continuous variables. The chi-square test was utilized for comparison of categorical variables. After the receiver operating characteristic (ROC) curve was plotted, the area under the curve (AUC) was calculated to evaluate the diagnostic efficacy of SOX9. For survival comparison between high and low SOX9 expression groups, the Kaplan–Meier method with the log-rank test was used. Univariate analysis and multivariate analysis were performed using the Cox regression model. Only variables with P≤0.10 in univariate analysis were included in multivariate analysis. All experiments were replicated at least three times. P<0.05 was considered statistically significant.
Results
Expression of SOX9 in HCC and adjacent normal tissue
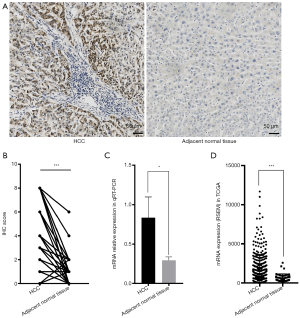
Representative photographs of IHC analysis of the HCC tissues and adjacent normal tissues were shown in Figure 1A. The statistical result of IHC analysis revealed that the expression of SOX9 in HCC was markedly higher than that in adjacent normal tissues (n=99 pairs, Figure 1B). The qRT-PCR results were consistent with the IHC analysis (n=8 pairs, Figure 1C). Additionally, higher SOX9 expression in HCC over adjacent normal tissues was also validated in the TCGA database including 374 HCC patients and 50 normal liver tissues (Figure 1D). In all, these findings strongly indicated that SOX9 is an oncogene in HCC.
Expression of SOX9 in CHCC and NCHCC
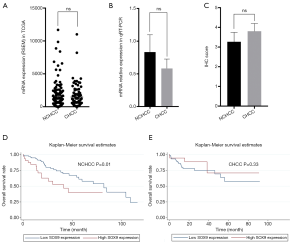
Using the data from the TCGA database, which included 79 CHCC and 133 NCHCC tissues, we found that CHCC and NCHCC showed no difference in the expression of SOX9 (Figure 2A). The qRT-PCR experiment, which included 8 pairs of CHCC and NCHCC tissues, also indicated that expression of SOX9 was not different between CHCC and NCHCC tissues (Figure 2B). Consistently, IHC analysis of 62 CHCC and 37 NCHCC tissues also showed no difference between CHCC and NCHCC (Figure 2C). In summary, these results indicate that CHCC and NCHCC are not different with respect to expression of SOX9.
Prognostic value of SOX9 in CHCC and NCHCC
Using data from TCGA, we evaluated the prognostic significance of SOX9 in NCHCC (Table 1) and CHCC (Table 2) by survival analysis. For NCHCC, univariate analysis indicated that high SOX9 expression, tumor T stage, N stage, and M stage were all risk factors for OS. The Kaplan-Meier survival curve showed a difference in OS between the high/low SOX9 expression groups in NCHCC (Figure 2D). Multivariate analysis showed that high SOX9 expression and M stage in tumors were independent risk factors of OS. On the other hand, for CHCC in TCGA, univariate analysis found that old age and vessels in the tumor were risk factors of OS. The Kaplan-Meier survival curve showed no difference in OS rates between the high/low SOX9 expression groups in CHCC (Figure 2E). Multivariate analysis showed that old age and vessels in the tumor were independent risk factors of OS. Overall, our findings revealed that SOX9 exhibits differential prognostic value between CHCC and NCHCC.
Table 1
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age (≥60) | 1.06 (0.57–1.95) | 0.86 | |||
| Gender (male) | 0.82 (0.45–1.49) | 0.52 | |||
| Family cancer history (yes) | 1.72 (0.95–3.13) | 0.07 | 1.78 (0.93–3.43) | 0.08 | |
| Serum AFP level (≥400) | 0.95 (0.43–2.10) | 0.90 | |||
| Child-Pugh classification (B/A) | 1.13 (0.39–3.22) | 0.83 | |||
| Pathologic stage (III + IV/I + II) | 1.72 (0.93–3.18) | 0.08 | 3.13 (0.34–28.67) | 0.31 | |
| T stage (T3 + T4 + Tx/T1 + T2) | 1.99 (1.09–3.63) | 0.03 | 0.46 (0.05–4.36) | 0.50 | |
| N stage (N1 + N2 + Nx/N0) | 2.15 (1.14–4.04) | 0.02 | 1.35 (0.53–3.48) | 0.53 | |
| M stage (M1 + Mx/M0) | 2.52 (1.33–4.78) | <0.01 | 2.95 (1.16–7.48) | 0.02 | |
| Vessels in tumor (Micro + Macro/none) | 1.12 (0.57–2.20) | 0.75 | |||
| Recurrence (yes) | 1.37 (0.75–2.50) | 0.37 | |||
| SOX9 (high expression) | 2.20 (1.16–4.16) | 0.02 | 2.23 (1.07–4.62) | 0.03 | |
NCHCC, noncirrhotic hepatocellular carcinoma; HR, hazard ratio; CI, confidence interval; AFP, α-fetoprotein; Micro, microvessels; Macro, macrovessels; SOX9, Sex-Determining Region Y-Box 9.
Table 2
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age (≥60) | 3.03 (1.15–7.97) | 0.02 | 3.14 (1.16–8.49) | 0.02 | |
| Gender (male) | 0.90 (0.30–2.73) | 0.85 | |||
| Family cancer history (yes) | 0.85 (0.27–2.67) | 0.78 | |||
| Serum AFP level (≥400) | 1.61 (0.52–4.94) | 0.41 | |||
| Child-Pugh classification (B/A) | 2.94 (0.82–10.60) | 0.10 | 4.04 (0.94–17.30) | 0.06 | |
| Pathologic stage (III + IV/I + II) | 2.42 (0.80–7.36) | 0.12 | |||
| T stage (T3 + T4 + Tx/T1 + T2) | 2.89 (0.95–8.78) | 0.06 | 1.60 (0.44–5.85) | 0.48 | |
| N stage (N1 + N2 + Nx/N0) | 1.08 (0.35–3.38) | 0.89 | |||
| M stage (M1 + Mx/M0) | 2.20 (0.77–6.30) | 0.14 | |||
| Vessels in tumor (Micro + Macro/none) | 3.57 (1.44–8.86) | <0.01 | 3.51 (1.28–9.62) | 0.02 | |
| Recurrence (yes) | 1.36 (0.52–3.60) | 0.53 | |||
| SOX9 (high expression) | 0.55 (0.16–1.89) | 0.34 | |||
CHCC, cirrhotic hepatocellular carcinoma; HR, hazard ratio; CI, confidence interval; AFP, α-fetoprotein; Micro, microvessels; Macro, macrovessels; SOX9, Sex-Determining Region Y-Box 9.
Correlation of SOX9 with clinicopathological traits in CHCC and NCHCC
Similarly, using data from TCGA, correlation analysis was conducted in NCHCC (Table S2) and CHCC (Table S3). In both CHCC and NCHCC, SOX9 expression was positively associated with serum AFP levels (P<0.05). However, in CHCC or NCHCC, other variables, including age, sex, family cancer history, pathological stage, T stage, N stage, M stage, recurrence, and vessels in the tumor showed no significant association with SOX9 expression (P>0.05).
Diagnostic value of SOX9 in CHCC and NCHCC
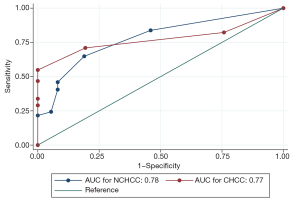
The correlation analysis in the TCGA database demonstrated that expression of SOX9 was positively related to serum AFP levels in both CHCC and NCHCC. The results suggested that SOX9 might have diagnostic value in identifying HCC, similar to the widely used serum AFP levels in clinical practice. To quantify and certify the diagnostic efficacy of SOX9, using clinicopathological data from our cohort (99 pairs of HCC tissues and adjacent normal tissues were included), we graphed ROC curves and calculated the AUC value (Figure 3). As a consequence, with the data from IHC analysis, the AUC of SOX9 in differentiating HCCs and adjacent tissues in CHCC and NCHCC was 0.77 (95% CI: 0.69–0.86) and 0.78 (95% CI: 0.68–0.88), respectively. The statistical test indicated that the AUCs for CHCC and NCHCC were not significantly different (P=0.95).
Discussion
After reviewing previous articles, it was found that expression of SOX9 was not limited to a particular tissue or cell type (13). It is primarily located in the nucleus and acts as a transcription factor to regulate the expression of many genes (14). SOX9 was previously found to participate in the processes of chondrogenesis, sex differentiation, heart development, tumor occurrence, and tumor progression. Malignant behaviors induced by ectopic expression of SOX9 have been identified in colorectal cancer, prostate cancer, pancreatic cancer, lung cancer, and melanoma (15-18).
In our study, expression of SOX9 was higher in HCC than in adjacent normal tissues. This differential expression was also reported in published studies using HCC tissues and cell lines (19,20). These results indicate that SOX9 should be classified as an oncogene in HCC. In our study, correlation analysis found that SOX9 expression in both CHCC and NCHCC was positively related to serum AFP levels. Therefore, we speculated that SOX9 might have diagnostic potential in HCC. We then used ROC curves and AUC to evaluate its diagnostic value. We found that the AUCs for NCHCC and CHCC were all higher than 0.75, indicating that the diagnostic ability of SOX9 is high and reliable.
Expression of ATAD2 (21), CTC-297N7.9 (22), ENST00000429227.1 (23), and PARPBP (24) in HCC tissues has been reported to be associated with serum AFP levels, and they were thought to be promising biomarkers beneficial for the molecular and pathological diagnosis of HCC. Therefore, based on these findings, we conclude that SOX9 may represent an indicator for the diagnosis of HCC patients. Because the AUCs were not different between CHCC and NCHCC, the diagnostic efficacy of SOX9 was thought to be comparable between CHCC and NCHCC. Previous studies have found many differences between CHCC and NCHCC, but they also identified some similarities, such as number and distribution of nodules (4). Therefore, CHCC might not be completely distinct from NCHCC with respect to clinicopathological traits or gene expression. In other words, they can also exhibit some similarities. Our study found, the form of expression and the role of SOX9 as a diagnostic marker are similarities between CHCC and NCHCC.
Our study found that CHCC and NCHCC showed no difference in SOX9 expression. However, this does not mean that SOX9 has the same prognostic value in both CHCC and NCHCC. Therefore, we conducted survival analysis to evaluate the prognostic value of SOX9 in CHCC and NCHCC. Kaplan-Meier analysis indicated that higher SOX9 expression was associated with reduced OS in NCHCC. However, in CHCC, no difference in OS was found between high and low SOX9 expression groups. In addition, univariate and multivariate analyses also revealed that high SOX9 expression was a risk factor for poor survival in NCHCC but not in CHCC. Taken together, the results of the survival analyses indicated that SOX9 may represent a prognostic indicator for NCHCC but not for CHCC. Therefore, with respect to prognosis, the value of SOX9 expression in CHCC and NCHCC appears to be different. Currently, proposed prognostic markers for CHCC or NCHCC are relatively limited. A previous study found that mir-149 was a prognostic marker in NCHCC but not in CHCC (25). Similarly, herein, we found that SOX9 is a prognostic marker for NCHCC but not for CHCC. These findings are helpful to guide prognostic evaluation in patients with NCHCC.
Furthermore, in both NCHCC and CHCC, we found that SOX9 expression was not associated with other traits, such as pathological stage, tumor recurrence, tumor T stage, N stage, and M stage. Two previous studies also suggested that there was no association between SOX9 expression and pathological stage, tumor T stage, N stage, or M stage in HCC (12, 26). Therefore, SOX9 expression might not be associated with common clinicopathological traits. Whether SOX9 is related to other clinicopathological traits still needs further exploration.
Our study also has some limitations. First, our study found that SOX9 is a prognostic marker for NCHCC. However, how it influences prognosis has not been clarified. Therefore, further efforts should be made to determine the molecular mechanisms of SOX9 in NCHCC. Second, although the role of SOX9 in HCC was explored using the TCGA database and our cohort, the sample size was still limited. Therefore, further study with larger numbers of tissue samples is still needed for verification of the current findings. Regardless of these shortcomings, our study certified the prognostic and diagnostic value of SOX9 in CHCC and NCHCC.
In conclusion, in CHCC and NCHCC, high SOX9 expression may represent a reliable diagnostic marker to differentiate HCC from normal liver tissues. SOX9 is thought to have a more important function in NCHCC than in CHCC because high SOX9 expression was a marker for poor prognosis only in NCHCC.
Acknowledgments
Funding: The study was funded by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tcr-20-3385
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-20-3385
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-20-3385). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics committee of the Second Affiliated Hospital of Chongqing Medical University (NO. 2020-329) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Younossi ZM, Koenig AB, Abdelatif D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73-84. [Crossref] [PubMed]
- Kanwal F, Kramer J, Asch SM, et al. Risk of Hepatocellular Cancer in HCV Patients Treated With Direct-Acting Antiviral Agents. Gastroenterology 2017;153:996-1005.e1. [Crossref] [PubMed]
- Conti F, Buonfiglioli F, Scuteri A, et al. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J Hepatol 2016;65:727-33. [Crossref] [PubMed]
- Schütte K, Schulz C, Poranzke J, et al. Characterization and prognosis of patients with hepatocellular carcinoma (HCC) in the non-cirrhotic liver. BMC Gastroenterol 2014;14:117. [Crossref] [PubMed]
- Núñez Martínez Ó, Matilla Peña A, Merino Rodríguez B, et al. Descriptive study of hepatocellular carcinoma in noncirrhotic liver. Gastroenterol Hepatol 2011;34:322-8. [PubMed]
- Giannini EG, Marenco S, Bruzzone L, et al. Hepatocellular carcinoma in patients without cirrhosis in Italy. Dig Liver Dis 2013;45:164-9. [Crossref] [PubMed]
- Zhang Y, Wang C, Xu H, et al. Hepatocellular carcinoma in the noncirrhotic liver: a literature review. Eur J Gastroenterol Hepatol 2019;31:743-8. [Crossref] [PubMed]
- Desai A, Sandhu S, Lai JP, et al. Hepatocellular carcinoma in non-cirrhotic liver: A comprehensive review. World J Hepatol 2019;11:1-18. [Crossref] [PubMed]
- Cai C, Wang H, He HH, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Invest 2013;123:1109-22. [Crossref] [PubMed]
- Wang H, He L, Ma F, et al. SOX9 regulates low density lipoprotein receptor-related protein 6 (LRP6) and T-cell factor 4 (TCF4) expression and Wnt/β-catenin activation in breast cancer. J Biol Chem 2013;288:6478-87. [Crossref] [PubMed]
- Zhang Y, Guo X, Xiong L, et al. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett 2012;586:4362-70. [Crossref] [PubMed]
- Liu C, Liu L, Chen X, et al. Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology 2016;64:117-29. [Crossref] [PubMed]
- Prévostel C, Blache P. The dose-dependent effect of SOX9 and its incidence in colorectal cancer. Eur J Cancer 2017;86:150-7. [Crossref] [PubMed]
- Matsushita M, Kitoh H, Kaneko H, et al. A novel SOX9 H169Q mutation in a family with overlapping phenotype of mild campomelic dysplasia and small patella syndrome. Am J Med Genet A 2013;161A:2528-34. [PubMed]
- Pritchett J, Athwal V, Roberts N, et al. Understanding the role of SOX9 in acquired diseases: lessons from development. Trends Mol Med 2011;17:166-74. [Crossref] [PubMed]
- Zhou CH, Ye LP, Ye SX, et al. Clinical significance of SOX9 in human non-small cell lung cancer progression and overall patient survival. J Exp Clin Cancer Res 2012;31:18. [Crossref] [PubMed]
- Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet 2009;25:19-29. [Crossref] [PubMed]
- Zhang S, Zhu C, Zhu L, et al. Oncogenicity of the transcription factor SOX8 in hepatocellular carcinoma. Med Oncol 2014;31:918. [Crossref] [PubMed]
- Liu Y, Zhang W, Liu K, et al. miR-138 suppresses cell proliferation and invasion by inhibiting SOX9 in hepatocellular carcinoma. Am J Transl Res 2016;8:2159-68. [PubMed]
- Leung CO, Mak WN, Kai AK, et al. Sox9 confers stemness properties in hepatocellular carcinoma through Frizzled-7 mediated Wnt/β-catenin signaling. Oncotarget 2016;7:29371-86. [Crossref] [PubMed]
- Meng X, Wang L, Zhu B, et al. Integrated Bioinformatics Analysis of the Clinical Value and Biological Function of ATAD2 in Hepatocellular Carcinoma. Biomed Res Int 2020;2020:8657468. [Crossref] [PubMed]
- Zhu S, Huang X, Zhang K, et al. Low expression of long noncoding RNA CTC-297N7.9 predicts poor prognosis in patients with hepatocellular carcinoma. Cancer Med 2019;8:7679-92. [Crossref] [PubMed]
- Zhao Y, Kong CQ, Ye JZ, et al. Upregulation of Long Non-Coding RNA ENST00000429227.1 Is Correlated with Poor Prognosis in Human Hepatocellular Carcinoma. Med Sci Monit 2019;25:6539-46. [Crossref] [PubMed]
- Yu B, Ding Y, Liao X, et al. Overexpression of PARPBP Correlates with Tumor Progression and Poor Prognosis in Hepatocellular Carcinoma. Dig Dis Sci 2019;64:2878-92. [Crossref] [PubMed]
- Mei Y, You Y, Xia J, et al. Identifying Differentially Expressed MicroRNAs Between Cirrhotic and Non-Cirrhotic Hepatocellular Carcinoma and Exploring Their Functions Using Bioinformatic Analysis. Cell Physiol Biochem 2018;48:1443-56. [Crossref] [PubMed]
- Richtig G, Aigelsreiter A, Schwarzenbacher D, et al. SOX9 is a proliferation and stem cell factor in hepatocellular carcinoma and possess widespread prognostic significance in different cancer types. PLoS One 2017;12:e0187814. [Crossref] [PubMed]


