
Prognostic significance of pyruvate kinase M2 expression in esophageal squamous cell carcinoma and its meta-analysis
Introduction
Esophageal cancer (EC) is among the most common gastrointestinal tract tumors. According to the most recent International Agency for Research on Cancer (IARC) report, 455,800 new EC cases (3% of all cancers) were reported and 400,200 deaths due to EC (5% of all cancer deaths) occurred worldwide in 2012 (1). China has one of the highest rates of EC in the world, with an average of about 150,000 deaths per year due to EC. Esophageal squamous cell carcinoma (ESCC) is the predominant type of EC in China (2). Clinical treatment methods include surgical treatment, radiotherapy, chemotherapy, endoscopic treatment, traditional Chinese medicine antitumor treatment, palliative treatment, and others. Although these traditional treatments have been used in clinical practice for many years, the prognosis of patients with ESCC has remained poor, with an overall 5-year survival rate of 18% (3). Therefore, new efficient prognostic markers need to be identified for risk estimation in ESCC patients.
Numerous oncological studies have focused on cancer metabolism due to the aberrant feature of energy production. Tumor cells acquire the vast majority of energy from glycolysis and lactic acid fermentation regardless of sufficient oxygen supply; this unique phenomenon is known as the Warburg effect, or aerobic glycolysis (4). Pyruvate kinase (PK) is one of the main rate-limiting enzymes in glycolysis. There are 2 isoenzymes, M-type and I-type, and the M-type has M1 and M2 isoforms (PKM1 and PKM2). The PKM1 and PKM2 isoforms are different in their expression and function (5). The PKM2 isoform is mainly expressed in differentiated tissues, such as those of the lung, adipose tissue, retina, and islet, and is also expressed in cells with a high nucleic acid synthesis rate, such as normal proliferating cells, embryonic cells, and tumor cells (6). There have been several findings which have alerted researchers to the potential role of PKM2 in tumorigenesis. Sizemore et al. (7) confirmed that the serine/threonine kinase ataxia telangiectasia-mutated (ATM) phosphorylates nuclear PKM2 at T328 following DNA damage, leading to the accumulation of PKM2 in the nucleus. Increased nuclear pT328-PKM2 level is associated with significantly worse survival in glioblastoma patients, so using PKM2-targeting strategies can not only disrupt cancer metabolism but can also inhibit an important mechanism of resistance to genotoxic therapies. For ESCC, PKM2 is overexpressed in ESCC tissues and cell lines (8), and PKM2 overexpression is associated with poor prognosis of ESCC (9-14).
However, the clinical significance of PKM2 expression remains controversial due to conflicting clinical evidence. Hence, we performed a meta-analysis to clarify the prognostic significance of PKM2 in ESCC and offer referential information for future clinical practice. We present the following article in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-442) (15).
Methods
Search strategy
The PubMed, Embase, Medline, and Cochrane Library databases were searched with the key phrases “esophageal squamous cell carcinoma” OR “oesophageal squamous cell carcinoma” OR “ESCC” AND “pyruvate kinase M2 OR PKM2”. Google Scholar and the latest conferences were also searched.
Selection criteria
The literature inclusion criteria were as follows: (I) patients in the original study histopathologically diagnosed with ESCC; (II) assessment of the expression of PKM2 with immunohistochemistry (IHC); (III) analysis of the associations of PKM2 with disease-specific survival (DSS), disease-free survival (DFS), progression-free survival (PFS), or overall survival (OS); and (IV) full text available. The exclusion criteria were as follows: Kaplan-Meier survival curve was not used for survival analysis in the original literature, or the data were incomplete and hazard ratio (HR) data could not be obtained.
Literature screening, data extraction, and quality evaluation
The retrieved literature was independently screened by two investigators (Q Zhang and S Zheng) by title and abstract for inclusion in the review. If the suitability of an article was uncertain, the full text was assessed. Disagreements were resolved through discussion or review by a third investigator (L Yang). The following information was extracted from the enrolled studies: general information (including the first author’s name, year of publication, article name, and publication details), number of participants, clinical features of participants, detection method, antibody, cutoff value of high and low expression of PKM2, high expression rate of PKM2, OS, estimate of the HRs, and 95% confidence intervals (CIs). If the HR was not directly provided in the original literature, a request was sent to the author via email. If the authors did not respond, the data were extracted from the article’s survival curve. The Newcastle-Ottawa Scale (NOS) was used by the two investigators to assess the quality of the original studies. Disagreements were resolved through discussion or reviewed by the third investigator. The NOS evaluates randomized, case-controlled, and cohort studies by evaluating population selection, comparability, exposure evaluation, or outcome evaluation. It contains 8 items, and the evaluation of literature quality is based on the semi-quantized principle of the star system, with a full score of 9. All studies with NOS scores of 6 or above are considered high quality.
Statistical analysis
Review Manager 5.3 (RevMan, Cochrane Collaborative, Oxford, UK) software was used in the meta-analysis. The software allowed the results to be presented graphically. Heterogeneity between studies was assessed by chi-squared (χ2)-based Q test and I2 tests, where I2 >50% or P<0.05 was considered to indicate significant heterogeneity. Publication bias was estimated using funnel plots and Egger’s test through the software Stata 14.0 (StataCorp, College Station, TX, USA). A P value <0.1 was considered to indicate statistically significant publication bias.
Results
The selection of research participants and their characteristics
The processes of identifying and selecting studies are presented in Figure 1. Through the preliminary search, a total of 153 studies were screened out. By reading the titles and abstracts of these articles, repetitive or irrelevant studies were excluded. After a detailed review of the remaining 16 studies that potentially met the inclusion criteria, 5 articles comprising 781 participants were finally included in the meta-analysis. The OS was documented in all studies. According to the NOS literature quality assessment scale, the quality score of all the studies was 7, indicating that the quality of the studies was relatively high (Table 1).
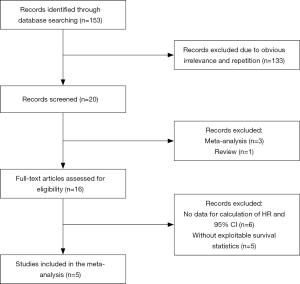
Table 1
| Study year | Country | Technology | Sample size | Age median | Gender (F/M) | PKM2 (L/H) | Follow-up (months) | Outcome | HR (95% CI) | Cutoff value | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fukuda 2015, (10) | Japan | IHC | 205 | NA | 30/175 | 101/104 | 47.9±43.4 | OS | 1.850 (1.200–2.780) | Score ≥6 | 7 |
| Li 2014, (11) | China | IHC | 141 | 60 | 54/87 | 82/59 | NA | OS | 1.214 (0.728–2.026) | Score ≥0.75 | 7 |
| Zhan 2013, (12) | China | IHC | 210 | NA | 48/162 | 43/167 | Overall 72.0 | OS | 1.748 (1.277–2.395) | Score ≥4 | 7 |
| Zhang 2013, (13) | China | IHC | 86 | 65 (41–81) | 22/64 | 24/62 | NA | OS | 2.358 (1.156–4.812) | Score ≥4 | 7 |
| Ma 2019, (14) | China | IHC | 139 | NA | 32/107 | 36/103 | NA | OS | 1.754 (1.070–2.876) | Score ≥3 | 7 |
CI, confidence interval; HR, hazard ratio; OR, odds ratio; IHC, immunohistochemistry; NOS, Newcastle-Ottawa Scale; NA, not available; OS, overall survival; PKM2, pyruvate kinase M2; F, female; M, male; H, high; L, low.
Association of PKM2 expression with OS
It has been reported that PKM2 is a key molecule in the metastasis of cancer and is overexpressed in various cancer tissues in comparison with paired normal adjacent tissue (NAT). However, there has been a lack of summary of different studies on PKM2 in ESCC to provide the reader with extensive information on the clinical impact of PKM2 in ESCC. Consequently, we meta-analyzed the expression of PKM2 in ESCC in the present study. Among the 5 included articles, different antibody manufacturers were used, and the dilution ratio was 1:100, except in the study of Zhang et al. (13), in which the ratio was 1:30. The IHC method used was either the EnVision (Agilent Technologies, Santa Clara, CA, USA) method or the streptavidin peroxidase (SP) method. The scoring systems mainly included staining intensity and the percentage of positive cells. The staining intensity of 5 articles was scored as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. The percentage of positive cells was scored slightly differently between the 5 articles (Table S1). Therefore, the definition of the cutoff value in the 5 articles was different. For Fukuda et al. (10), Zhan et al. (12), and Zhang et al. (13), the cutoff value was defined as the median value multiplied by the intensity score and the percentage of positive cells score. For Ma et al. (14), the cutoff value was the median of the staining intensity score plus the percentage of positive cells score. For Li et al. (11), the cutoff value was defined by combining the weighted score generated by the multiplication of the intensity score and the percentage of positive cells score and statistical analysis. All of them qualified PKM2 expression as “low” when the immunoreactive score (IRS) was less than the cutoff value and “high” when it was higher than the cutoff value.
We found that analysis of PKM2 expression in all cases showed that overexpression of PKM2 was associated with poor prognosis in patients with ESCC (HR =1.72, 95% CI: 1.41–2.09; P<0.01; Figure 2A).
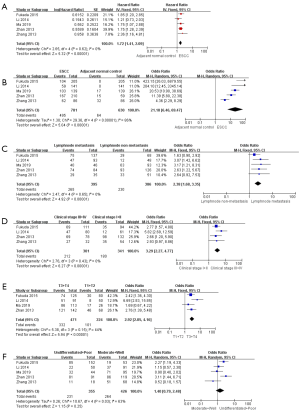
Association of PKM2 expression with clinicopathological features
Positivity of PKM2 and its overexpression were significantly different between ESCC and its paired normal controls (OR =21.18, 95% CI: 6.46–69.47; P<0.01) and significantly different between lymph node metastasis and non-metastasis (OR =2.38, 95% CI: 1.68–3.35; P<0.01; Figure 2B,C). The positivity and overexpression of PKM2 were significantly associated with clinical stage I–II and clinical stage III–IV (OR =3.29, 95% CI: 2.27–4.77; P<0.01) and significantly associated with T classification (OR =2.92, 95% CI: 2.05–4.16; P<0.01; Figure 2D,E). However, PKM2 positivity and overexpression were not significantly associated with tumor differentiation (OR =1.40, 95% CI: 0.79–2.48; P=0.25; Figure 2F). Together, these data indicate that PKM2 overexpression could significantly correlate with lymph node metastasis, clinical stage, and T classification in tissues of ESCC.
Heterogeneity analysis
Heterogeneity among the studies was analyzed by χ2 test and I2 test, and heterogeneity was found in correlation analysis of PKM2 expression between ESCC and NAT (P<0.05; I2 =86%) and tumor differentiation (P<0.05; I2 =63%). Therefore, the random effects model was used to analyze PKM2 expression between ESCC and NAT and tumor differentiation, and the fixed effects model was used for other correlation analyses.
Publication bias
Begg’s funnel plots were created to assess the publication bias of the articles. The peaks of the funnel plots did not reveal any evidence of obvious asymmetry (Figure 3). For the accuracy of the results, Egger’s test was used to further evaluate publication bias, with the results showing no publication bias among the studies (P>0.05; Table 2).
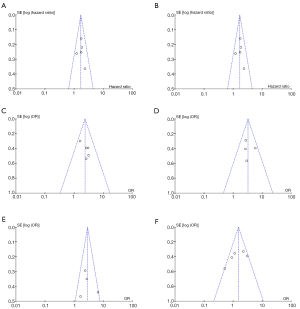
Table 2
| Comparison | t | P value | 95% CI |
|---|---|---|---|
| ESCC and NAT | 1.65 | 0.197 | 2.838–8.966 |
| Lymph node metastasis | 2.20 | 0.115 | 1.206–6.622 |
| Clinical stage | 0.26 | 0.820 | 10.255–11.571 |
| T classification | 0.38 | 0.740 | 16.753–20.005 |
| Tumor differentiation | 1.47 | 0.237 | 18.106–6.649 |
ESCC, esophageal squamous cell carcinoma; NAT, normal adjacent tissue; CI, confidence interval.
Discussion
It is currently understood that PKM2 plays an important role in the glucose metabolism of malignant tumors, and the PKM2-mediated Warburg effect can provide sufficient energy and a large number of metabolic intermediates for the rapid proliferation of tumor cells (16). The reason is that under the regulation of various factors, PKM2 can switch between the highly active tetramer and the less active dimer (17), with the tetramer having PK activity, and the dimer having protein kinase activity (18). The activity of PKM2 is regulated by a variety of posttranslational modifications, such as phosphorylation, acetylation, small ubiquitin-like modifier (SUMO)-ylation, hydroxylation, and oxidation, which prefer the formation of dimer PKM2 in tumor cells (18,19). Mutations of PKM2 can also change its activity (20). A study showed that PKM2 activity is downregulated by the oxidation of C358 by reactive oxygen species (ROS) or hypoxia, leading to switching of the flux of glucose into the pentose phosphate pathway and glycolytic biosynthesis to generate nicotinamide adenine dinucleotide phosphate (NADPH) for ROS detoxification and tumor progression (21). Cytoplasmic PKM2 is a stable tetramer form, and nuclear PKM2 is a dimer form that plays the role of protein kinase and uses phosphoenolpyruvate (PEP) as a phosphate donor (22). In the nucleus, STAT3 is phosphorylated at tyrosine 705 by PKM2.
The above studies have suggested that PKM2 figures prominently in the emergence and development of malignant tumors. However, the prognostic value of PKM2 in ESCC has not yet been determined. Therefore, the relationship between PKM2 expression and ESCC prognosis and clinicopathological parameters was systematically evaluated and summarized in this meta-analysis. The results showed that PKM2 was expressed differently in ESCC and paired NAT, and the prognostic analysis suggested that PKM2 overexpression was related to the poor prognosis of ESCC. It was also suggested that PKM2 overexpression correlates with lymph node metastasis, clinical stage, and T classification in ESCC tissues.
Similarly, some studies have suggested that PKM2 can be used as a prognostic marker for pancreatic ductal adenocarcinoma (PDAC), breast cancer, hepatocellular carcinoma (HCC), and gallbladder carcinoma (23-25). However, the prognostic value of PKM2 remains controversial. We performed this meta-analysis to provide a more comprehensive and direct understanding of whether PKM2 can be used as a prognostic marker for ESCC. In addition, as lymph node metastasis is the most important prognostic factor in ESCC (26), accurate nodal staging is crucial for the treatment of ESCC (27). Some studies report PKM2 expression to not be associated with lymph node metastasis in ESCC (10,12,13). Therefore, a meta-analysis combining the results of several studies enabled a more comprehensive overview. Our study showed that PKM2 overexpression correlates with lymph node metastasis of ESCC, suggesting that PKM2 may be a molecular target for lymph node metastasis of ESCC. It is also controversial whether PKM2 is associated with tumor differentiation in ESCC. Our results showed that PKM2 was not associated with tumor differentiation.
An interesting finding was that strong PKM2 expression significantly correlated with poor response to chemotherapy. Fukuda et al. (10) showed that strong PKM2 expression significantly correlated with decreased OS in patients who received neoadjuvant chemotherapy followed by surgery, and PKM2 expression was not affected by the neoadjuvant chemotherapy. Therefore, the therapeutic value of PKM2 should be systematically assessed. Liu et al. (28) reported that the PKM2 inhibitor shikonin inhibited proliferation and glycolysis and induced cell apoptosis in HCC cells. James et al. (29) reported that PKM2 inhibitor shikonin reduced PDAC cell proliferation, cell migration, and induced cell death. Tang et al. (30) reported that shikonin enhances sensitization of gefitinib against wild-type epidermal growth factor receptor (EGFR) non-small cell lung cancer via inhibition of the PKM2/STAT3/cyclinD1 signal pathway. Another study reported that shikonin has a significant antitumor effect in EC by regulating the HIF1α/PKM2 signal pathway (31). Considering the complex function of PKM2 in cell biology, measures that inhibit or silence PKM2 possibly cause a wide range of effects in the human body, especially in patients who are chemotherapy resistant.
Despite producing valuable findings, there were a few limitations to our study. First, although we did not find any obvious evidence for publication bias from funnel plots and Egger’s tests, this meta-analysis was based on formally published articles with principally positive results. Hence, there was a potential publication bias that might have lowered the accuracy and validity of the results. Second, due to some relatively small sample-sized studies and some missing information, the quality of the included studies was not uniform.
In summary, the present study suggests that PKM2 is crucial for the development of ESCC and that PKM2 overexpression is associated with poor prognosis of ESCC and correlates with lymph node metastasis, clinical stage, and T classification. There is potential for PKM2 as a potential prognostic biomarker and therapeutic target for ESCC.
Conclusions
The present study demonstrated that PKM2 plays a crucial role in ESCC. The level of PKM2 is significantly associated with ESCC prognosis and tumor-node-metastasis staging. Additional research is needed to investigate how PKM2 promotes metastasis during ESCC carcinogenesis.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-442
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-442
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-442). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yang X, Chen X, Zhuang M, et al. Smoking and alcohol drinking in relation to the risk of esophageal squamous cell carcinoma: A population-based case-control study in China. Sci Rep 2017;7:17249. [Crossref] [PubMed]
- Doi T, Piha-Paul S, Jalal S, et al. Safety and Antitumor Activity of the Anti-Programmed Death-1 Antibody Pembrolizumab in Patients with Advanced Esophageal Carcinoma. J Clin Oncol 2018;36:61-7. [Crossref] [PubMed]
- Fu J, Xiong Z, Huang C, et al. Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus. J Biol Chem 2019;294:327-40. [Crossref] [PubMed]
- Yao G, Yin J, Wang Q, et al. Glypican-3 Enhances Reprogramming of Glucose Metabolism in Liver Cancer Cells. Biomed Res Int 2019;2019:2560650. [Crossref] [PubMed]
- Hsu MC, Hung WC. Pyruvate kinase M2 fuels multiple aspects of cancer cells: from cellular metabolism, transcriptional regulation to extracellular signaling. Mol Cancer 2018;17:35. [Crossref] [PubMed]
- Xu D, Liang J, Lin J, et al. PKM2: A Potential Regulator of Rheumatoid Arthritis via Glycolytic and Non-Glycolytic Pathways. Front Immunol 2019;10:2919. [Crossref] [PubMed]
- Sizemore ST, Zhang M, Cho JH, et al. Pyruvate kinase M2 regulates homologous recombination-mediated DNA double-strand break repair. Cell Res 2018;28:1090-102. [Crossref] [PubMed]
- Li S, Huang P, Gan J, et al. Dihydroartemisinin represses esophageal cancer glycolysis by down-regulating pyruvate kinase M2. Eur J Pharmacol 2019;854:232-9. [Crossref] [PubMed]
- Liu Q, Liang M, Liu T, et al. M2 isoform of pyruvate kinase (PKM2) is upregulated in Kazakh's ESCC and promotes proliferation and migration of ESCC cells. Tumour Biol 2016;37:2665-72. [Crossref] [PubMed]
- Fukuda S, Miyata H, Miyazaki Y, et al. Pyruvate Kinase M2 Modulates Esophageal Squamous Cell Carcinoma Chemotherapy Response by Regulating the Pentose Phosphate Pathway. Ann Surg Oncol 2015;22:S1461-8. [Crossref] [PubMed]
- Li W, Xu Z, Hong J, et al. Expression patterns of three regulation enzymes in glycolysis in esophageal squamous cell carcinoma: association with survival. Med Oncol 2014;31:118. [Crossref] [PubMed]
- Zhan C, Shi Y, Lu C, et al. Pyruvate kinase M2 is highly correlated with the differentiation and the prognosis of esophageal squamous cell cancer. Dis Esophagus 2013;26:746-53. [Crossref] [PubMed]
- Zhang X, He C, He C, et al. Nuclear PKM2 expression predicts poor prognosis in patients with esophageal squamous cell carcinoma. Pathol Res Pract 2013;209:510-5. [Crossref] [PubMed]
- Ma R, Liu Q, Zheng S, et al. PKM2-regulated STAT3 promotes esophageal squamous cell carcinoma progression via TGF-β1-induced EMT. J Cell Biochem 2019;120:11539-50. [Crossref]
- Liberati A, Altman D, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 2009;339:b2700. [Crossref] [PubMed]
- Li Z, Yang P, Li Z. The multifaceted regulation and functions of PKM2 in tumor progression. Biochim Biophys Acta 2014;1846:285-96. [PubMed]
- van Niekerk G, Engelbrecht A. Role of PKM2 in directing the metabolic fate of glucose in cancer: a potential therapeutic target. Cell Oncol (Dordr) 2018;41:343-51. [Crossref] [PubMed]
- Zahra K, Dey T. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front Oncol 2020;10:159. [Crossref] [PubMed]
- Wang Y, Liu J, Jin X, et al. O-GlcNAcylation destabilizes the active tetrameric PKM2 to promote the Warburg effect. Proc Natl Acad Sci U S A 2017;114:13732-7. [Crossref] [PubMed]
- Liu VM, Howell AJ, Hosios AM, et al. Cancer-associated mutations in human pyruvate kinase M2 impair enzyme activity. FEBS Lett 2020;594:646-64. [Crossref] [PubMed]
- Anastasiou D, Poulogiannis G, Asara J, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011;334:1278-83. [Crossref] [PubMed]
- Xiaoyu H, Yiru Y, Shuisheng S, et al. The mTOR Pathway Regulates PKM2 to Affect Glycolysis in Esophageal Squamous Cell Carcinoma. Technol Cancer Res Treat 2018;17:1533033818780063. [Crossref] [PubMed]
- Hu H, Tu W, Chen Y, et al. The combination of PKM2 overexpression and M2 macrophages infiltration confers a poor prognosis for PDAC patients. J Cancer 2020;11:2022-31. [Crossref] [PubMed]
- Yang Y, Wu K, Liu Y, et al. Prognostic significance of metabolic enzyme pyruvate kinase M2 in breast cancer: A meta-analysis. Medicine (Baltimore) 2017;96:e8690. [Crossref] [PubMed]
- Zhu H, Luo H, Zhu X, et al. Pyruvate kinase M2 (PKM2) expression correlates with prognosis in solid cancers: a meta-analysis. Oncotarget 2017;8:1628-40. [Crossref] [PubMed]
- Wu L, Yang X, Cao W, et al. Multiple Level CT Radiomics Features Preoperatively Predict Lymph Node Metastasis in Esophageal Cancer: A Multicentre Retrospective Study. Front Oncol 2020;9:1548. [Crossref] [PubMed]
- Li B, Li N, Liu S, et al. Does [18F] fluorodeoxyglucose-positron emission tomography/computed tomography have a role in cervical nodal staging for esophageal squamous cell carcinoma? J Thorac Cardiovasc Surg 2020;160:544-50. [Crossref] [PubMed]
- Liu T, Li S, Wu L, et al. Experimental Study of Hepatocellular Carcinoma Treatment by Shikonin Through Regulating PKM2. J Hepatocell Carcinoma 2020;7:19-31. [Crossref] [PubMed]
- James AD, Richardson DA, Oh IW, et al. Cutting off the fuel supply to calcium pumps in pancreatic cancer cells: role of pyruvate kinase-M2 (PKM2). Br J Cancer 2020;122:266-78. [Crossref] [PubMed]
- Tang JC, Ren YG, Zhao J, et al. Shikonin enhances sensitization of gefitinib against wild-type EGFR non-small cell lung cancer via inhibition PKM2/stat3/cyclinD1 signal pathway. Life Sci 2018;204:71-7. [Crossref] [PubMed]
- Tang JC, Zhao J, Long F, et al. Efficacy of Shikonin against Esophageal Cancer Cells and its possible mechanisms in vitro and in vivo. J Cancer 2018;9:32-40. [Crossref] [PubMed]


