Effect of LHX2 gene methylation level and its function on radiotherapy of cervical cancer
Introduction
Cervical cancer is the most common malignancy of the female reproductive system, with nearly 500,000 new cases worldwide and more than 50% cases resulting in death (1,2) each year. There are many factors that induce cervical cancer, including human papillomavirus (HPV), lifestyle habits, unhealthy behaviors, family inheritance, smoking, multiple births, oral contraceptives, and so on. Among them, HPV is the most important pathogenic factor for cervical cancer. At present, HPV vaccination and early diagnosis are considered effective strategies to prevent this disease (3), but the survival rate from cervical cancer was still low, especially in developing countries.
Methylation of DNA is one of the most common DNA epigenetic modifications in eukaryotic cells. It refers to the process of covalency binding of methyl groups provided by S-adenosine methionine (SAM) to specific bases under the action of DNA methylation transferase. Aberrant DNA methylation has been extensively studied in recent years in the tumor context. Hypermethylation of tumor suppressor genes, that results in silencing, or hypomethylation of oncogenes, that can activate these genes, are important mechanisms in the tumor development and progression. Many DNA methylation-based assays have described many methylated genes as promising markers for the management of HPV-positive women, such as SOX1, PAX1, JAM3, EPB41L3, CADM1, and MAL (4-6), which can be applied for the detection of advanced cervical squamous intraepithelial neoplasia 2/3 (CIN2/3) lesions (7). While alterations in DNA methylation has been documented in multiple studies for these genes, an important gap in our knowledge exist regarding the functional consequences of the altered gene expression in the development and progression of cervical cancer.
The National Comprehensive Cancer Network (NCCN) Guidelines provide recommendations for cervical cancer treatment, stipulating that surgery is the predominant intervention for cervical cancer treatment, combined with radiotherapy and adjuvant chemotherapy (7). In clinical treatment, radiotherapy and chemotherapy are both known to cause acute and late toxicity (8). In addition, increasing number of evidences suggest that changes in cancer methylome is associated with the acquisition of drug resistance as well (9). It has been reported that radiation reduced the expression of the key enzymes in methylation formation, such as DNA (cytosine-5)-methyltransferase 1 (DNMT1), DNMT3A, and DNMT3B, and further reduced genomic methylation (10,11). These changes lead to increased expression of tumor antigen genes. The mutation rate of the K-ras (Kirsten rat sarcoma viral oncogene homolog) gene in cancer tissues before radiotherapy was 44%, which reduced to 12% after radiotherapy. The mutation rate of the MGMR (O-6-Methylguanine-DNA Methyltransferase) gene in cancer tissues before radiotherapy was 36%, and it dropped to 8% after radiotherapy (12). In statistical analysis, radiotherapy increased the relative survival rates, but also showed poorer prognosis in younger cervical cancer patients (13). In the clinical process of diagnosis and treatment, the tumor size and International Federation of Obstetrics and Gynecology (FIGO) stage may efficiently serve as markers for responsiveness to radiotherapy, but tumor cells resistance to radiation obscure a major challenge.. Therefore, identifying new molecular targets is crucial to further explore the mechanism of cervical cancer and assess the radiation sensitivity.
Members of the LHX gene family perform important functions in different tissues and biological functions, especially in tissue-specific differentiation and body size formation. There have been 9 members (LHX 1-LHX 9) identified in the LHX family in mammals. Some LHX family members have been shown to be closely associated with cancer. Transgenic expression of LHX2 promotes vascular maturation, primary tumor growth, tumor cell infiltration, and metastasis during breast cancer oncogenesis (14). In hepatocellular carcinoma, LHX3 is a late prognostic biomarker and a metastatic oncogene (15,16). The LHX4 gene may inhibit tumorigenesis by downregulating the expression of alpha-fetoprotein during the development of liver cancer (17). Epigenetic changes of the LHX9 gene contribute to the invasion and migration of glioma cells (10). Based on the throughput data analysis of methylation sequencing, it was found that LHX9 genes were highly methylated in cervical cancer tissues, which was significantly decreased after radiotherapy treatment.
The level of gene methylation in cells is an important factor involved in the development of tumors, and radiation therapy is an important means to change the DNA methylation state of cells in the body. In vivo radiotherapy experiments in mice have demonstrated that methylation changes simultaneously exhibit radiation dose effects (11). The expression of DNMT1, DNMT3A, and DNMT3B, the key enzymes involved in methylation at irradiated sites, was decreased by radiation, causing extensive demethylation reactions in the genome and resulting in histone H2Ax-Ser139 phosphorylation of γH2Ax (an early reaction of DNA breakage) and demethylation of histone H4-Lys20 (18). These changes promote the expression of tumor antigen genes. In another study, the K-ras and MGMR gene mutation rates were 44% and 36.0%, respectively, before radiotherapy in cancer tissues, and were 12% and 8% after radiotherapy, respectively, indicating that radiotherapy could improve the control of local tumors and was of great significance for surgical treatment (19). Studies have shown that genome-wide breast cancer cells with low methylation have lower radiosensitivity than hypermethylated cell lines, and activation of methyltransferase by SAM causes hypermethylation and high radiosensitivity (10). However, changes in methylation levels have a gene-specific effect on radiosensitivity. Hypermethylation of specific genes (Axin, ATM, MGMT, and TIMP3) or hypomethylation (SerpinB5 and S100A6) alter gene expression (20) and disrupt biological functions, eventually increasing radiosensitivity. Combined with the analysis of the above two aspects, we hypothesized that radiation directly caused DNA methylation changes and promoted the expression of tumor suppressor genes to inhibit the occurrence and development of tumors. Therefore, the study of methylation of key genes in cervical cancer represents a new research direction and may provide new ideas for the treatment of cervical cancer.
In this study, cervical cancer cells (siHA and C33A) were selected as the research object to detect the expression and methylation level of LHX2. Meanwhile, demethylation, methylation, and LHX2 overexpression were used to treat the cells, and radiotherapy was performed on the cells to detect the changes of LHX2 expression and methylation level in the cells after radiotherapy. We studied the transcriptome, proteome analysis, cell cycle, cell apoptosis, DNA damage, and repair detection to explore the molecular mechanism of LHX2 in affecting the function of cervical cancer. It is hoped that LHX2 may become a candidate gene for the investigation of molecular mechanism of cervical cancer.
We believe that our approach, including high-throughput candidate identification, combined with gain- and loss-of-function in vitro validations represent an improved approach that will not only identify putative markers, but also provide insight into the molecular mechanisms how the identified biomarkers alter the pathogenesis of cervical cancer.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-739).
Methods
Ethics
This study was approved by the Ethics Committee of Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China (YKD201902089); all participants provided written informed consent. All experiments were designed and performed following the relevant guidelines. Cervical cancer tissue samples were obtained from patients with a diagnostic report in Department of Radiation Oncology, Inner Mongolia Cancer Hospital & Affiliated People’s Hospital of Inner Mongolia Medical University. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Study participants, tissue sample preparation and collection
All tissue samples were collected after surgical operation. Cervical cancer tissue samples before and after radiotherapy from three different patients were also collected. Meanwhile, cervical cancer tissue samples as well as peripheral blood samples were collected from 47 different patients. All tissues samples were frozen in liquid nitrogen. The serum was obtained by centrifugation and frozen in liquid nitrogen. All tissue samples were used for DNA and RNA extraction. Cervical cancer patients ranged from 18 to 70 years old. The FIGO staging levels were from IIB to VA.
Analysis of the LHX2 promotor methylation
The bisulfite sequencing PCR (BSP) primers were designed using Methyl Primer Express v 1.0 (Thermo Fisher Scientific, Waltham, MA USA).
The polymerase chain reaction (PCR) was performed in 50 µL with 8 µL of 10× PCR buffer (Mg2+), 4 µL of a deoxynucleotide triphosphate (dNTP) mixture (each 2.5 mM), 0.5 µL Taq polymerase (5 U/L), 1 µL of F and R primers (10 µM), and 5–10 ng environmental DNA as template. The following PCR program was employed: 95 °C for 5 min; 30 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 25 s; 72 °C for 7 min; and hold at 4 °C. The positive clones were sent to Sangon Biotech (Shanghai) Co., Ltd. for sequencing. The methylation rate of each sample was calculated using the following formula:
Methylation rate of each sample (%) = C/(*Nc*n)100%
Where C indicates the number of methylation sites in each sample, N indicates the sample number, Nc indicates the number of cloning sites, and n indicates the number of CpG sites.
The methylation rate of each group of samples was calculated as follows:
Methylation rate of each group (%) = Methylation rate of each sample in the group/N.
RNA extraction and first-strand cDNA synthesis and quantitative real-time PCR validation
The RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA synthesis was made using cDNA kit, Roche Cat# 04379012001.
The primers were designed using Prime Primer 5 and synthesized by the Beijing Genomics Institute (Table 1), and β-Actin (ACT3) served as an internal control. Quantitative real-time PCR (qRT-PCR) was conducted using SYBR Green dye (LightCycle® 480 SYBR Green I master, Roche). The selected genes were verified using the LightCycle® 480 Real-Time PCR System (Roche). The following PCR program was employed: 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 20 s. The dissolution curve was assessed at 72–95 °C. The heating rate was 0.5 °C/10 s. Fold change expression of genes were calculated using 2−△△Ct method.
Table 1
| Number | Pathologic types | FIGO staging | LHX2 | |
|---|---|---|---|---|
| Gene expression | Promoter methylation level | |||
| 800***286 | Squamous | IVB | 0.24 | 0.48 |
| 800***672 | Squamous | IIIC | 0.30 | 0.46 |
| 800***441 | Squamous | IIA | 1.00 | 0.40 |
| 800***734 | Squamous | IIIC | 0.25 | 0.48 |
| 800***887 | Squamous | IIIC | 0.35 | 0.56 |
| 800***752 | Squamous | IIIC | 0.20 | 0.45 |
| 800***001 | Squamous | IIIC | 2.26 | 0.31 |
| 800***006 | Squamous | IIIC | 0.20 | 0.46 |
| 800***978 | Squamous | IIB | 1.00 | 0.39 |
| 800***084 | Squamous | IIIC | 0.80 | 0.42 |
| 800***657 | Squamous | IIIC | 0.40 | 0.52 |
| 800***531 | Squamous | IIB | 1.75 | 0.36 |
| 800***869 | Squamous | IIIB | 0.43 | 0.53 |
| 800***518 | Squamous | IIA | 0.28 | 0.43 |
| 800***921 | Squamous | IIIC | 0.15 | 0.45 |
| 800***647 | Squamous | IIIC | 0.29 | 0.43 |
| 800***814 | Squamous | IIIB | 1.00 | 0.44 |
| 800***418 | Squamous | IIIC | 4.49 | 0.21 |
| 800***033 | Squamous | IIIC | 0.84 | 0.42 |
| 800***164 | Squamous | IIB | 0.93 | 0.40 |
| 800***252 | Squamous | IIIC | 1.00 | 0.40 |
| 800***268 | Squamous | IIB | 1.97 | 0.33 |
| 800***084 | Squamous | IIIC | 0.46 | 0.51 |
| 800***538 | Squamous | IIIC | 1.39 | 0.38 |
| 800***844 | Squamous | IIIC | 0.15 | 0.47 |
| 800***243 | Squamous | IIIC | 53.20 | 0.32 |
| 800***918 | Squamous | IIA | 1.00 | 0.40 |
| 800***645 | Squamous | IIB | 138.99 | 0.16 |
| 800***791 | Squamous | IIIC | 20.01 | 0.18 |
| 800***394 | Squamous | IIB | 111.77 | 0.17 |
| 800***860 | Squamous | IIIC | 94.73 | 0.30 |
| 800***847 | Squamous | IIIC | 52.93 | 0.36 |
| 800***248 | Squamous | IIB | 3.03 | 0.39 |
| 800***524 | Squamous | IIB | 15.62 | 0.24 |
| 800***713 | Squamous | IIB | 18.99 | 0.19 |
| 800***329 | Squamous | IVB | 1.00 | 0.41 |
| 800***773 | Squamous | IIIC | 2.92 | 0.35 |
| 800***920 | Squamous | IIIC | 20.55 | 0.19 |
| 800***991 | Squamous | IIIC | 1.38 | 0.39 |
| 800***878 | Squamous | IIIC | 1.00 | 0.41 |
| 800***937 | Squamous | IIIC | 0.06 | 0.45 |
| 800***597 | Squamous | IIIC | 29.61 | 0.32 |
| 800***887 | Squamous | IIIC | 0.43 | 0.46 |
| 800***755 | Squamous | IIB | 1.65 | 0.36 |
| 800***897 | Squamous | IIB | 13.74 | 0.25 |
| 800***097 | Other | IIIC | 0.79 | 0.42 |
| 800***246 | Squamous | IIIC | 0.91 | 0.42 |
FIGO, International Federation of Obstetrics and Gynecology.
Immunohistochemistry of cervical cancer samples
Tissues were incubated overnight in formalin for fixation and embedded in paraffin. 4 µm thick sections were used. Serum albumin was used for blocking non-specific sites.. Sections were incubated overnight with primary antibodies at 4 °C, followed by a short incubation with fluorescent conjugated secondary antibodies for 30 min at room temperature. 3’3-diaminobenzidine HCl (DAB) incubation was done for 1 min.
Cell culture
The siHA and C33A cell lines were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM, HyClone Laboratories, Inc., Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) and 1% P/S at 37 °C in an incubator with 5% CO2.
Radiation treatment
Cervical cancer tissues and cell lines (siHA and C33A) were inoculated in culture flasks at a density of 1×105. After 24 h of adherent culture, X-ray irradiation was performed. To confirm the 6 MV X-ray effect on two types of cells, we exposed cells to 5 different doses of radiation (2, 4, 6, 8, and 10 Gy). After 24 h, cells exposed to 2 and 4 Gy exhibited normal cell morphology and cell numbers. Exposure to 6–10 Gy of irradiation cased cells to enter apoptosis, so these cells were not used in subsequent experiments. An experimental dose of 2 Gy was chosen. After 48 h of radiation treatment, the culture medium was discarded, and the cells were digested with trypsin and collected. The cells were washed twice with phosphate buffered saline (PBS) and then collected.
Plasmid and transfections
Small interfering RNA (siRNA) to knockdown LHX2 and the negative control were purchased from GenePharma (Shanghai, China). Transient transfections were performed with the help of GenePharma (Shanghai, China) following the instructions of manufacturer’s. For stable transfections, overexpression plasmid (pcDNA 3.1+) was employed to construct the recombinant plasmids pcDNA 3.1-LHX2 (inserted with LHX2). The cells were then incubated in G418 containing medium, and G418-resistant cells were obtained approximately 35 days after transfection.
Cell viability assay, migration and invasiveness assays
Cell viability was assessed using cell Counting Kit-8 (CCK-8). Cells (2×103/well) were seeded into a 96-well plate followed by overnight incubation. The cells were washed with PBS after medium was removed. The cells were then incubated in the presence of medium (90 µL) and CCK-8 (10 µL) and incubated for 2 h at 37 °C; the optical density (OD) was measured at 450 nm using microplate reader.
We used Boyden chambers with pore size of 8 µm for the migration assay (BD Biosciences, Becton, Dickinson and Co., Franklin Lakes, NJ, USA). 5×104 cells 200 µL medium containing 5×104 cells and 0.7 ml of DMEM were placed in the upper chamber, and lower chamber. Following 24 h of incubation, using cotton swab cells were removed from the upper side of the filter, and fixed with methanol for 10 min followed by crystal violet stain. The remaining cells under the filter were counted. For invasion assay, same conditions were except chambers were coated with Matrigel (BD Bioscience, San Jose, CA, USA).
Western blot
Protein lysates were prepared in lysis buffer, on ice and incubated for 30 min, followed by centrifugation at 15,000 rpm for 15 min at 4 °C. For immunoprecipitation (IP). the precipitated proteins were collected using A+G beads, followed by washing and eluting using Laemmli buffer (Thermo Fisher Scientific, Waltham, MA, USA), and finally subjected to western blotting. Same amount of protein was loaded on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to nitrocellulose membrane (Millipore, Bedford, MA, USA). The membranes were probed overnight at 4 °C with primary antibodies purchased from Abnova (Beijing, China) and Cell Signaling Technology. Blots were then incubated with HRP conjugated secondary antibodies and developed with ECL reagents (Invitrogen, USA).
Statistical analysis
The results were expressed as mean ± standard error of the mean (SEM). Statistical analyses were performed using SPSS version 18.0 software (IBM Corp., Chicago, IL, USA). The student’s t-test was used to compare two different groups, while the ANOVA test was used for variances of three or more groups. Spearman’s rank correlation was used for the analysis of correlation between two gene expressions.
Results
The expression and methylation level of LHX2 gene
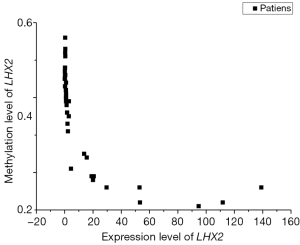
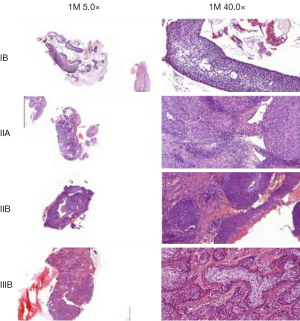
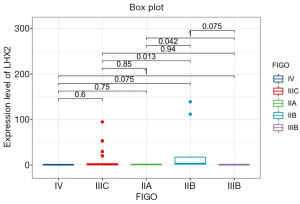
The correlation analysis of gene expression and methylation level of LHX2 genes in cervical cancer tissues with staging promoter from 47 participants were diagnosed with cervical squamous cell carcinoma. According to the FIGO guidelines, the types of cervical cancer were divided into six stages, including 3 IIA type, 11 IIB type, 2 IIIB type, 29 IIIC type and 2 IV type. FIGO stages was confirmed with HE stains results of patients (Figure 1). After sequencing and bioinformatics analysis, the methylation levels of LHX2 were detected by BSP. The results showed that the upstream sequence was at high methylation in all tested participants. The methylation level of LHX2 was 16–56% in patients (P<0.05) (Table 1 and Figure 2). Moreover, the pyrosequencing results of samples from participants showed an upward trend of the methylation level of LHX2 with the change of FIGO stage (Figure 3).
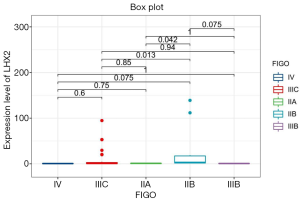
The correlation analysis of gene expression and methylation level of LHX2 gene in cervical cancer tissues with staging promoter from 47 patients were diagnosed with cervical squamous cell carcinoma. In this study, the expression level and methylation level of LHX2 was detected in 47 specimens. The correlation between LHX2 expression level and methylation level with FIGO staging were analyzed using SPSS software. The results showed that the expression level of LHX2 gene was significantly correlated with IIB and IIIC stage (P<0.05) (Figure 4). The IIB and IIA stage was different significantly correlated with the expression level of LHX2 gene (P<0.05). Meanwhile, the methylation level of LHX2 gene promoter was significantly correlated with IIB and IIIB stage (P<0.05), IIA and IIB stage (P<0.05), IIB and IIIC stage (P<0.05), IIB and IV stage (P<0.05) (Figure 5).
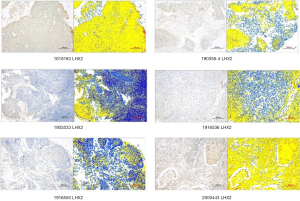
Immunohistochemistry analysis of LHX2 protein
In this study, 37 cervical cancer tissues were treated with paraffin embedding. The immunohistochemistry (IHC) analysis showed that LHX2 protein expression was satisfactory (Table 2). The characteristics of LHX2 protein expression were consistent with the course and stage of the disease in all samples (Figure 6). The expression level of the LHX2 proteins were downregulated with the development of disease stage.
Table 2
| Names | Positive area % | Mean density | Area density | H-score |
|---|---|---|---|---|
| 1618183 LHX2 100 | 81.16 | 0.04 | 0.03 | 96.57 |
| 190358-4 LHX2 100 | 16.72 | 0.05 | 0.01 | 33.88 |
| 1905533 LHX2 100 | 16.03 | 0.05 | 0.01 | 18.47 |
| 1916536 LHX2 100 | 31.70 | 0.03 | 0.01 | 43.90 |
| 1916550 LHX2 100 | 17.18 | 0.04 | 0.01 | 22.26 |
| 192410-3 LHX2 100 | 7.20 | 0.05 | 0.00 | 7.90 |
| 192773-6 LHX2 100 | 8.13 | 0.05 | 0.00 | 8.83 |
| 1928288 LHX2 100 | 44.38 | 0.04 | 0.02 | 58.26 |
| 2000443 LHX2 100 | 79.28 | 0.05 | 0.04 | 107.54 |
| 20172013 LHX2 100 | 29.92 | 0.04 | 0.01 | 37.37 |
| 162519-2 LHX2 100 | 41.08 | 0.04 | 0.02 | 47.17 |
| 170432-4 LHX2 100 | 42.94 | 0.05 | 0.02 | 79.70 |
| 171692- LHX2 100 | 60.06 | 0.06 | 0.03 | 85.64 |
| 171692- LHX2 100 | 27.37 | 0.05 | 0.01 | 35.20 |
| 175778- LHX2 100 | 51.37 | 0.05 | 0.03 | 72.06 |
| 177070-3 LHX2 100 | 18.06 | 0.06 | 0.01 | 26.08 |
| 181702- LHX2 100 | 42.29 | 0.04 | 0.02 | 50.27 |
| 182327- LHX2 100 | 43.30 | 0.05 | 0.02 | 54.65 |
| 183895- LHX2 100 | 32.25 | 0.05 | 0.02 | 45.17 |
| 184049- LHX2 100 | 77.26 | 0.04 | 0.03 | 94.67 |
| 188005- LHX2 100 | 43.86 | 0.04 | 0.02 | 52.61 |
| 188106- LHX2 100 | 63.11 | 0.04 | 0.03 | 84.91 |
| 188726- LHX2 100 | 55.22 | 0.04 | 0.02 | 72.80 |
| 189352-2 LHX2 200 | 10.62 | 0.04 | 0.00 | 18.84 |
| 189362-2 LHX2 100 | 30.44 | 0.04 | 0.01 | 35.96 |
| 189504-3 LHX2 100 | 8.28 | 0.05 | 0.00 | 10.81 |
| 189801-2 LHX2 200 | 9.33 | 0.04 | 0.00 | 10.69 |
| 1900401-2 LHX2 100 | 51.95 | 0.04 | 0.02 | 73.34 |
| 1901586-2 LHX2 100 | 54.24 | 0.06 | 0.03 | 72.83 |
| 1902444-3 LHX2 100 | 24.81 | 0.04 | 0.01 | 43.41 |
| 1903438-1 LHX2 100 | 45.09 | 0.05 | 0.02 | 54.42 |
| 1908060- LHX2 100 | 39.50 | 0.07 | 0.03 | 68.78 |
| 1910137- LHX2 100 | 29.22 | 0.04 | 0.01 | 39.74 |
| 1910513- LHX2 100 | 8.79 | 0.05 | 0.00 | 9.71 |
| 2001134- LHX2 100 | 28.11 | 0.03 | 0.01 | 52.74 |
| 2002132-2 LHX2 100 | 35.10 | 0.04 | 0.01 | 39.36 |
| 173741-3 LHX2 100 | 36.37 | 0.05 | 0.02 | 49.00 |
| HLC LHX2 100 | 41.60 | 0.05 | 0.02 | 45.50 |
Enzyme-linked immunosorbent assay of LHX2 protein expression in serum of patients
The LHX2 protein in the serum of cervical cancer patients was tested by enzyme-linked immunosorbent assay (ELISA) to find a key gene which would predict the prognosis of radiotherapy by peripheral blood test in vitro (Table 3). Although the messenger RNA (mRNA) relative expression and methylation was significant in cancer tissues, but there was no significant change in serum during radiation treatment. The results showed that the expression of LHX2 protein in serum was not significantly correlated with early, middle, and late radiotherapy treatment (P<0.05). The reason may be directly related to the function and characteristics of LHX2 protein. Generally speaking, most of the proteins that can enter the peripheral blood are secreted proteins, and they are ideally the indicators for in vitro detection, and their concentration should also be able to reach the detection line of ELISA experimental technology.
Table 3
| Number | Before treatment | Being treatment | After treatment |
|---|---|---|---|
| 800***286 | 0.516 | 0.443 | – |
| 800***672 | 0.5 | 0.543 | 0.469 |
| 800***441 | 0.505 | 0.544 | 0.538 |
| 800***734 | 0.526 | – | 0.524 |
| 800***502 | 0.536 | 0.611 | 0.571 |
| 800***752 | 0.493 | 0.524 | – |
| 800***001 | 0.541 | 0.539 | 0.443 |
| 800***006 | 0.497 | 0.561 | 0.602 |
| 800***978 | 0.499 | 0.567 | 0.556 |
| 800***084 | – | 0.464 | 0.463 |
| 800***657 | 0.445 | 0.521 | – |
| 800***531 | 0.533 | 0.52 | 0.513 |
| 800***869 | 0.522 | 0.495 | 0.557 |
| 800***300 | 0.533 | 0.532 | 0.537 |
| 800***588 | 0.632 | 0.604 | 0.618 |
| 800***518 | 0.603 | – | 0.584 |
| 800***921 | 0.497 | 0.548 | – |
| 800***347 | 0.482 | – | – |
| 800***647 | 0.519 | 0.611 | 0.627 |
| 800***814 | 0.628 | 0.589 | 0.526 |
| 800***418 | 0.589 | – | 0.504 |
| 800***161 | 0.415 | 0.426 | 0.432 |
ELISA, enzyme-linked immunosorbent assay.
LHX2 gene methylation levels in siHA and C33A cells
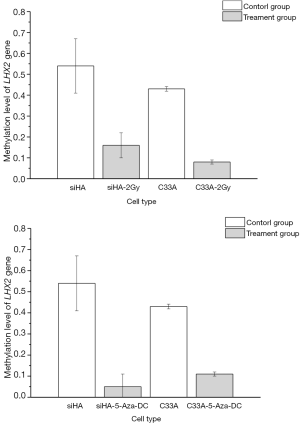
To identify the methylation status of the LHX2 genes in siHA and C33A squamous cell carcinoma cells. It was found that the methylation level of LHX2 was high in the cervical squamous cell line. In siHA cells, the methylation rates in the promoter region of LHX2 gene were 54%. In the C33A cell line, methylation level of LHX2 was similar to those in the siHA cell line. The methylation level of the LHX2 promoter was 43%. Statistical analysis showed that LHX2 was highly methylated in squamous cell lines, and significantly higher than in normal cells and tissues (Figure 2).
Gene expression characteristics of LHX2 in squamous carcinoma siHA and C33A cells after radiation
LHX2 methylation levels in cervical cancer cells after radiation treatment
After radiation treatment, the BSP-PCR detection results showed that methylation level of LHX2 changes significantly in the cells of siHA-2-Gy, the methylation level of LHX2 promoter region methylation rate reduced to 16%. In C33A-2Gy cell line, the methylation level of LHX2 promoter region methylation rate reduced to 8%. The methylation rate decreased significantly (P<0.05). The results are shown in Figure 2.
Gene expression characteristics of LHX2 in squamous cell carcinoma after radiation treatment
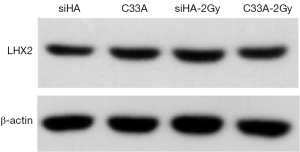
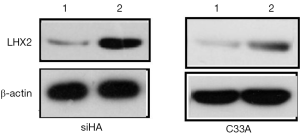
After treatment with 2Gy radiation rays, the expression levels of LHX2 in C33A-2Gy were 3.5 times of those cervical tissues (cells without radiation treatment). The relative expressions of LHX2 in siHA-2Gy cells were increased by 2.3 times. Meanwhile, the same result was detected in the expression level of LHX2 protein, the WB detection result showed the expression level of LHX2 protein was up regulated in the in squamous cell carcinoma after radiation (Figure 7).
Gene expression characteristics in squamous carcinoma siHA and C33A cells after treatment with 5-Aza-DC
Effects of 5-Aza-DC on methylation levels of LHX2 genes
After 5-Aza-DC treatment, the BSP-PCR detection results showed that methylation level of LHX2. LHX2 changes significantly in the cells of siHA-5-Aza-DC, the methylation level of LHX2 reduced to 5%. In C33A-2Gy cell line, the methylation level of LHX2 reduced to 11%. The methylation rate decreased significantly (P<0.05). The results were shown in Table 4. After 5-Aza-DC treatment, the methylation levels of the two types of cells were not consistent, and the corresponding gene changes in siHA cells were more obvious, while C33A cells were less sensitive to 5-Aza-DC at 5 µM.
Table 4
| Group | Methylation rate of LHX2 (%) |
|---|---|
| C33A | 43 |
| C33A-5-Aza-dC | 11 |
| siHA | 54 |
| siHA-5-Aza-dC | 5 |
Effects of 5-Aza-DC on expression level of LHX2 genes
The relative expression levels of LHX2 gene in siHA and C33A carcinoma cells were measured by qRT-PCR. The results showed that the expression levels of LHX2 were up regulated in both treatment cell lines compared with control groups (Figure 8). The expression levels of LHX2 in C33A-5-Aza-dC was 14.2 times of those without 5-Aza-DC treatment (Figure 8 and Table 5). Meanwhile, the same result was detected in the expression level of LHX2 protein, the WB detection result showed the expression level of LHX2 protein was up regulated in the in squamous cell carcinoma after treatment with 5-Aza-DC (Figure 7).
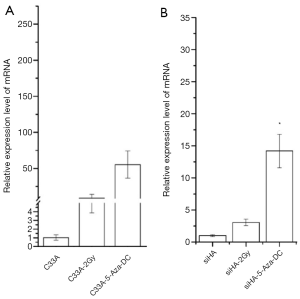
Table 5
| Group | Expression level of LHX2 | P value |
|---|---|---|
| C33A | 1.03±0.32 | |
| C33A-2Gy | 8.98±5.11 | <0.05 |
| C33A-5-Aza-dC | 55.34±18.83 | <0.01 |
| siHA | 1.01±0.15 | |
| siHA-2Gy | 3.06±0.51 | <0.05 |
| siHA-5-Aza-dC | 14.20±2.60 | <0.05 |
Effects of 5-Aza-dC and radiation on the cell invasion and migration of siHA and C33A cells
In order to confirm the effect of 5-Aza-dC and radiotherapy on invasion and migration of cervical cancer cells, 5-Aza-dC and radiation were applied to treat cells. The invasion results showed that the number of transmembrane cells in the siHA group, siHA-5-Aza-dC group, and siHA-2Gy group were 153±4.39, 116.67±3.06, and 125.37±11.37, respectively. The number of transmembrane cells in the C33A group, C33A-5-Aza-dC and C33A-2Gy group were 176.67±10.78, 155.33±11.37, and 153.33±7.37, respectively. The statistical analysis results indicated that the number of transmembrane cells decreased after 2Gy radiotherapy for both types of cervical squamous cell carcinoma cells, and the difference was statistically significant (P<0.05) (Table 6).
Table 6
| Group | Cell number |
|---|---|
| siHA | 153±4.39 |
| siHA-5-Aza-dC | 116.67±3.06 |
| siHA-2Gy | 125.37±11.37 |
| C33A | 176.67±10.78 |
| C33A-5-Aza-dC | 155.33±11.37 |
| C33A-2Gy | 153.33±7.37 |
In the migration experiment (Table 7), the number of transferred cells were 296.67±15.26, 209.33±27.2274, and 182.66±14.57 in the siHA group, siHA-5-Aza-dC group, and siHA-2Gy group, respectively. In the C33A group, C33A-5-Aza-dC group, and C33A-2Gy group, the number of transferred cells were 229.33±15.01, 181.67±11.15, and 143±9.64, respectively. The results of statistical analysis indicated that the number of transmembrane metastatic cells decreased after 5-Aza-dC and 2 Gy radiotherapy for both types of cervical squamous cell carcinoma cells, and the differences were statistically significant (P<0.05), demonstrating that demethylation and radiotherapy inhibit the metastasis of cancer cells (Table 8).
Table 7
| Group and treatment | Cell number |
|---|---|
| siHA | 296.67±15.26 |
| siHA-5-Aza-dC | 209.33±27.2274 |
| siHA-2Gy | 182.66±14.57 |
| C33A | 229.33±15.01 |
| C33A-5-Aza-dC | 181.67±11.15 |
| C33A-2Gy | 143± 9.64 |
Table 8
| Group and treatment | Cell number |
|---|---|
| siHA | 258.66±18.58 |
| siHA-LHX2-OV | 317±6.21 |
| C33A | 268±10.44 |
| C33A-LHX2-OV | 312.33±10.78 |
Overexpression of LHX2
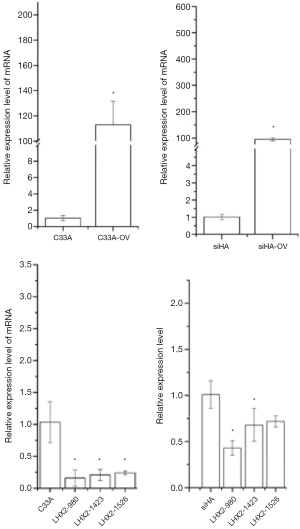
In the overexpression excrement, PCDNA-3.1 was used as the overexpression vector. The LHX2-overexpressed siHA and C33A cervical cancer cell lines (SiHa-LHX2-OV and C33A-LHX2-OV) were obtained by liposome transfection and G418 screening. The mRNA expression levels of LHX2 were 94 and 112 times more than that of the control group (Figures 9), but the difference of LHX2 methylation level was not significant between the overexpression group and control group. The western blot assay also confirmed the increased expression of the target gene (Figure 10). It was shown that our target gene overexpression cell line had been successfully constructed.
Effects of LHX2 overexpression on cell invasion and migration ability
Transwell invasion function of the overexpressed cell lines (siHA-LHX2-OV) was detected. The result shown that the number of transmembrane cells in siHA group and siHA-LHX2-OV group were 148.33±14.57 and 218±19.15. The number of transmembrane cells in C33A group and C33A-LHX2-OV group were 202±7.21 and 240±20.55. The overexpression of LHX2 in siHA and C33A cells resulted that Matrigel matrix was corroded, and the number of transferred cells were decreased. The difference was statistically significant (P<0.05) (Table 9).
Table 9
| Group and treatment | Cell number |
|---|---|
| siHA | 148.33±14.57 |
| siHA-LHX2-OV | 218±19.15 |
| C33A | 202±7.21 |
| C33A-LHX2-OV | 240±20.55 |
In the migration experiment, the number of transferred cells were 258.66±18.58 and 317±6.21 in the siHA group and siHA-LHX2-OV group. In the C33A group and C33A-LHX2-OV group, the number of transferred cells were 268±10.44 and 312.33±10.78. The results of statistical analysis indicated that the number of transmembrane metastatic cells decreased after LHX2 overexpression. The differences were statistically significant (P<0.05) (Table 8).
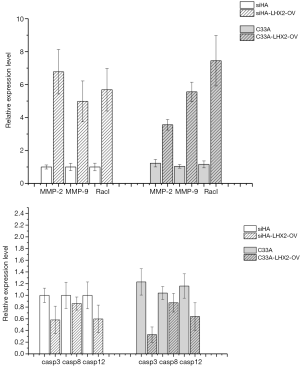
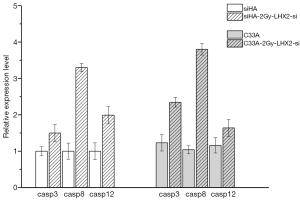
The Caspase-3, Caspase-8, and Caspase-12 genes were detected by qRT-PCR analysis to confirm the effects of overexpression of LHX2 on apoptosis-related proteins in cervical cancer cells. After overexpression, the expression of Caspase-3, Caspase-8, and Caspase-12 proteins were down-regulated. However, the expression of matrix metalloproteinase-2 (MMP-2), MMP-9, and RacI invasion-related genes were increased (Figure 11).
Effect of siRNA interference suppression on LHX2 in cells
In order to inhibit the increase of LHX2 expression induced by radiotherapy, siRNA interference was applied. The siRNA was transfected into siHA-LHX2-OV and C33A-LHX2-OV using liposomes. There were 3 kinds of siRNA transfected into siHA-LHX2-OV and C33A-LHX2-OV, including LHX2-980, LHX2-1423, and LHX2-1526. At 48 hours after transfection, the expression level of LHX2 was detected by RT-PCR (Figure 9). The LHX2-980 had a silencing efficiency of more than 80%, thus it was chosen for application in the siRNA interference experiment. Meanwhile, the same result was detected in the expression level of LHX2 protein, the WB detection result showed the expression level of LHX2 protein was silenced by siRNA-980 in the in squamous cell carcinoma after radiation (Figure 12).
After identifying the highest efficiency site, siRNA was transfected into cells with 2bGy radiation treatment for 24 h (siHA-LHX2-2Gy-si). After 48 h, the proliferation, metastasis, and apoptosis-related proteins of cells were detected.
The proliferation rate of radiotherapy treatment cell decreased after the LHX2 gene was silenced
Due to the timeliness of RNA silencing effect, in this study the proliferation results were detected at 48 h after transfection. The results showed that compared with siHA and C33A cells after irradiation, the cell proliferation rate was slightly up-regulated or down-regulated after silencing of the LHX2 gene, with no significant difference ranging from 8% to 15%. The results of the two kinds of cells were consistent, and the effect on C33A growth effect was smaller. It may be that the effects of radiation-induced and RNA silencing could cancel each other out. This resulted in no significant difference in gene expression.
In the invasion experiment, the speed of migration was not significantly different between the LHX2-2Gy-si group and control group. The number of invasions cells were 166.33±12.74 and 176±7 in the siHA-2Gy group and siHA-LHX2-2Gy-si group, respectively. In the C33A-2Gy group and C33A-LHX2-2Gy-si group, the number of invasions cells were 125.667±6.5 and 119.33±4.16. The results of statistical analysis indicated that the number of transmembrane metastatic cells increased after LHX2 silencing (Table 10).
Table 10
| Group and treatment | Cell number |
|---|---|
| siHA-2Gy | 166.33±12.74 |
| siHA-LHX2-2Gy-si | 176±7 |
| C33A-2Gy | 125.667±6.5 |
| C33A-LHX2-si | 119.33±4.16 |
In the migration experiment, the number of transferred cells were 116±10.45 and 120±7.6 in the siHA-2Gy and siHA-LHX2-2Gy-si. In C33A and C33A-LHX2-si group, the number of transferred cells were 98.33±9.5 and 109.76±6.52, respectively. The results of statistical analysis indicated that the number of transmembrane metastatic cells decreased after LHX2 overexpression. The differences were statistically significant (P<0.05) (Table 11).
Table 11
| Group and treatment | Cell number |
|---|---|
| siHA-2Gy | 116±10.45 |
| siHA-LHX2-2Gy-si | 120±7.6 |
| C33A-2Gy | 98.33±9.5 |
| C33A-LHX2-si | 109.76±6.52 |
The apoptosis of Radiotherapy treatment cell genes was inhibited after the expression of LHX2 being silenced
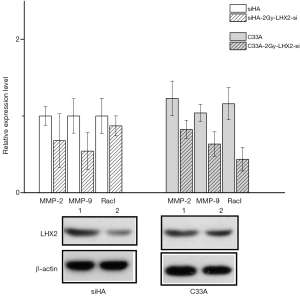
The Caspase-3, Caspase-8, and Caspase-12 genes were detected by qRT-PCR analysis to confirm the effects of siRNA of LHX2 on apoptosis-related proteins in cervical cancer cells. After siRNA, the expression of Caspase-3, Caspase-8, and Caspase-12 proteins were up-regulated. However, the expression of MMP-2, MMP-9, and RacI invasion-related genes were decreased (Figure 13).
Discussion
Cervical cancer is a malignant tumor, its morbidity and mortality account for the first and second highest among gynecological tumors in China, respectively. Radiotherapy is an important method for the treatment of cervical cancer, especially for patients with advanced or postoperative recurrence of cervical cancer, with good results. Although radiotherapy has an ideal therapeutic effect on cervical cancer, there are still some problems in the course of treatment, such as reduced radiation resistance and sensitivity and lack of sensitivity predictors. The methylation of DNA is an important epigenetic pattern that plays an important role in the structure and function of genomes. Methods for the detection of DNA methylation can be mainly divided into two categories: local methylation detection and genome-wide methylation detection. With the development of high-throughput sequencing, DNA methylation studies have moved into genome-wide methylation studies. Methylation-specific PCR (MSP), BSP, and other conventional methylation detection methods are limited to a few sites or local site research, and are low flux, time-consuming, and laborious. Combining high-throughput sequencing with BSP, DNA methylation studies can target a large number of sites, cover the entire genome, and provide the methylation levels of different functional regions simultaneously (21).
Romero-Garcia et al. has recently published a very thorough review on how DNA methylation can contribute to solid tumor resistance (9). For instance, Baharudin et al. performed DNA methylation profiling, and showed that patients of colorectal cancer (CRC) with 5-FU treatment significantly enhanced differentially methylated genes in CRC group compared to control (22). Another study by Guo et al. has found similarly critical role for DNA methylation in human lung adenocarcinoma in regard to resistant to cisplatin (23). Altogether, these findings along with other ongoing investigations, strongly implicate the important role of DNA methylation in radiotherapy.
DNA methylation has been previously studied in cervical cancer patients. A recent study has reported more than 100 human genes as possible methylation biomarkers of cervical cancer (24). Multiple panels containing several candidate host genes have been tested in different studies. Kong et al. recently reported that methylation of EPB41L3 and JAM3 is a feasible screening method for CIN2+ (25), showing that methylation testing achieved a remarkable success rate of 98.4%. Nevertheless, confirmatory studies and additional biomarkers are needed to develop even more reliable diagnostic pipelines.
Integration of gene expression and methylation profiles can greatly advance our understanding on the molecular mechanism driving cervical cancer progression. A recent study from Xu et al. provides a great example how such approach can be utilized (26). Xu et al. 2019 identified cervical cancer-specific DNA methylation markers. This study identified four markers specific methylation markers associated with cervical cancer. Databases sources like Gene Expression Omnibus (GEO) database or other sources are further required to confirm such results.
Combined with the difference in degree of methylation level and correlation enrichment analysis, we were able to determine which genes had significantly reduced methylation level after radiotherapy, which was speculated to be closely related to the pathogenesis of cervical cancer and the mechanism of radiotherapy, such as MYC, C10orf11, LHX1, LHX2, LHX5, and LHX9. The transcription factor MYC is closely associated with human malignancies. The c-myc protein can promote cells from the GL phase to S phase, promote cell proliferation and differentiation, and promote cell apoptosis (27,28). The results showed that c-myc plays a key role in the development of cervical lesions, so it may possibly be developed as a biomarker for the treatment of cervical lesions and subsequent decision-making, and c-myc is considered to be a very important biomarker (29). Studies have shown that the ANXA8 and C10ORF11 genes are candidate genes affecting lean body weight variation, which provides a new theoretical basis for the understanding of sarcopenia (30). In the clinical treatment, the radiotherapy effect of D and M patients is obvious, but the treatment effect of M patients is not obvious. At the same time, the methylation level of LHX5 and LHX9 genes was significantly decreased in cervical cancer tissue before and after radiotherapy in D and M patients. Therefore, this study paid special attention to these three genes.
We found that the protein products of the ISL-1 gene are similar to those of the MEC-3 (31) and LIN-11 (32,33) genes in that they have a cysteine-rich pattern upstream of the homologous domain. Therefore, the pattern containing similar structures is called LIM. The amino acid sequences encoded by LIM pattern color are called LIM domains, and the homeobox genes containing LIM pattern color are called LIM-homeobox genes. The LIM domain is also a sequence containing 55 amino acid residues that is very conserved at histidine and cysteine sites, whereas it is very poorly conserved between LIM domains of different genes or between different LIM domains of the same gene. The homeotypic domain downstream of LIM domain is well conserved. The LIM domain is a double zinc finger structure that can bind Zn2+ (34,35), because zinc finger proteins bind to DNA and regulate gene activity. In vitro experiments on MEC-3 and ISL-1 proteins showed that the binding of homologous domains to DNA in the proteins was regulated by the LIM domain (36).
As transcription factors, the LHX family of genes play an important role in signal transduction, cell differentiation, tissue-specific differentiation, and body size formation during the development of vertebrates and invertebrates (37). A total of 9 LHX members, named LHX1-9, have been found in mammals. The distribution of LHX1 is mainly throughout the central nervous system, but it has been confirmed by experiments that the forebrain and midbrain of mouse head are completely missing LHX1 (38), and it is speculated that LHX1 may act on the transcription of other genes to regulate the cell differentiation of these structures. The distribution of LHX2 is in multiple systems and it plays an important role. Mice with LHX2-/- have abnormal hippocampal primordia and cerebral cortex development, and their ocular primordia have been shown to stop developing prematurely (15). The role of LHX5 is important in regulating the morphological development of hippocampal structure. During gastrula formation, LHX5 was expressed in the hypothalamus of the diencephalon of mice. The function of LHX9 is similar to that of LHX2 in the nervous system, and LHX9 is also specifically expressed in the gonads of mice. Sperm stem cells of LHX9-/- mice can migrate, but somatic stem cells cannot regenerate, leading to abnormal gonadal development in these mice. It is speculated that under normal chromosomal conditions, the mutation of LHX9 may be closely related to abnormal gonadal development (14).
In addition to their role in tissue differentiation, the LHX family of genes is also closely associated with cancer. The gene LHX3 is a late prognostic biomarker and a metastatic oncogene in hepatocellular carcinoma (39). The gene LHX4 may inhibit the occurrence of tumors by down-regulating the expression of alpha-fetoprotein during the development of liver cancer (40). Studies have shown that LHX2 is an oncogene for a variety of tumors, and the transgenic expression of LHX2 promotes vascular maturation, primary tumor growth, tumor cell infiltration, and metastasis during the carcinogenesis of breast cancer (41). The LHX2 gene promotes the growth and metastasis of nasopharyngeal carcinoma by regulating the Wnt signaling pathway (42) and is abnormally expressed in lung cancer and pancreatic duct carcinoma (17). In the study of osteosarcoma, LHX2 promotes the occurrence of malignant tumor phenotypes by activating the mTOR pathway and inhibiting autophagy (43).
While our study has identified DNA methylation of LHX2 gene as an important and novel biomarker in cervical cancer radiotherapy, it is important to emphasize that our findings, and in general any epigenetic biomarkers need to be considered within the context of differential diagnostic situations, and this is particularly true for cervical cancer biomarkers (24,25).
Acknowledgments
Funding: This work was funded by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-739
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-739
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-739). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China (YKD201902089). All participants provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Wanqing C, Kexin S, Rongshou Z, et al. Cancer incidence and mort ality in China, 2014. ChinJ Cancer Res 2018;30:1-12. [Crossref]
- Crosbie EJ, Einstein MH, Franceschi S, et al. Human papillomavirus and cervical cancer. Lancet 2013;382:889-99. [Crossref] [PubMed]
- Lai HC, Lin YW, Huang RL, et al. Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer 2010;116:4266-74. [Crossref] [PubMed]
- van Leeuwen RW, Oštrbenk A, Poljak M, et al. DNA methylation markers as a triage test for identification of cervical lesions in a high risk human papillomavirus positive screening cohort. Int J Cancer 2019;144:746-54. [Crossref] [PubMed]
- Zummeren MV, Kremer WW, Leeman A, et al. HPV E4 expression and DNA hypermethylation of CADM1, MAL, and miR124-2 genes in cervical cancer and precursor lesions. Mod Pathol 2018;31:1842-50. [Crossref] [PubMed]
- Koh WJ, Abu-Rustum NR, Bean S, et al. Cervical Cancer, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2019;17:64-84. [Crossref] [PubMed]
- Green JA, Kirwan JM, Tierney JF, et al. Survival and recurrence after concomitant chemotherapy and radiotherapy for cancer of the uterine cervix: a systematic review and meta-analysis. Lancet 2001;358:781-6. [Crossref] [PubMed]
- Romero-Garcia S, Prado-Garcia H, Carlos-Reyes A. Role of DNA Methylation in the Resistance to Therapy in Solid Tumors. Front Oncol 2020;10:1152. [Crossref] [PubMed]
- Koturbash I, Baker M, Loree J, et al. Epigenetic dysregulation underlies radiation-induced transgenerational genome instability in vivo. Int J Radiat Oncol Biol Phys 2006;66:327-30. [Crossref] [PubMed]
- Pogribny I, Koturbash I, Tryndyak V, et al. Fractionated low-dose radiation exposure leads to accumulation of DNA damage and profound alterations in DNA and histone methylation in the murine thymus. Mol Cancer Res 2005;3:553-61. [Crossref] [PubMed]
- Ni SF. Detection and clinic Study of K-ras Mutations and MGMT Methylation in Rectal Cancer Undergone Preoperative Radiotherapy. Soochow University, 2006.
- Yagi A, Ueda Y, Kakuda M, et al. Epidemiologic and Clinical Analysis of Cervical Cancer Using Data from the Population-Based Osaka Cancer Registry. Cancer Res 2019;79:1252-9. [Crossref] [PubMed]
- Kuzmanov A, Hopfer U, Marti P, et al. LIM-homeobox gene 2 promotes tumor growth and metastasis by inducing autocrine and paracrine PDGF-B signaling. Mol Oncol 2014;8:401-16. [Crossref] [PubMed]
- Huang B, Tian ZF, Li LF, et al. LHX3 is an advanced-stage prognostic biomarker and metastatic oncogene in hepatocellular carcinoma. Cancer Biomark 2019;26:31-9. [Crossref] [PubMed]
- Hung TM, Hu RH, Ho CM, et al. Downregulation of alpha-fetoprotein expression by LHX4: a critical role in hepatocarcinogenesis. Carcinogenesis 2011;32:1815-23. [Crossref] [PubMed]
- Vladimirova V, Mikeska T, Waha A, et al. Aberrant methylation and reduced expression of LHX9 in malignant gliomas of childhood. Neoplasia 2009;11:700-11. [Crossref] [PubMed]
- Zhou LR, Cheng M, Qian LT. Advances in DNA methylation status and radiosensitivity of tumor cells. Chinese Journal of Radiation Oncology 2017;26:364-7.
- Barros-Silva D, Marques CJ, Henrique R, et al. Profiling DNA Methylation Based on Next-Generation Sequencing Approaches: New Insights and Clinical Applications. Genes (Basel) 2018;9:429. [Crossref] [PubMed]
- Jeong M, Guzman AG, Goodell MA. Genome-Wide Analysis of DNA Methylation in Hematopoietic Cells: DNA Methylation Analysis by WGBS. Methods Mol Biol 2017;1633:137-49. [Crossref] [PubMed]
- Ren R, Horton JR, Zhang X, et al. Detecting and interpreting DNA methylation marks. Curr Opin Struct Biol 2018;53:88-99. [Crossref] [PubMed]
- Baharudin R, Ab Mutalib NS, Othman SN, et al. Identification of Predictive DNA Methylation Biomarkers for Chemotherapy Response in Colorectal Cancer. Front Pharmacol 2017;8:47. [Crossref] [PubMed]
- Guo R, Wu G, Li H, et al. Promoter methylation profiles between human lung adenocarcinoma multidrug resistant A549/cisplatin (A549/DDP) cells and its progenitor A549 cells. Biol Pharm Bull 2013;36:1310-6. [Crossref] [PubMed]
- Lorincz AT. Virtues and Weaknesses of DNA Methylation as a Test for Cervical Cancer Prevention. Acta Cytol 2016;60:501-12. [Crossref] [PubMed]
- Kong L, Wang L, Wang Z, et al. DNA methylation for cervical cancer screening: a training set in China. Clin Epigenetics 2020;12:91. [Crossref] [PubMed]
- Xu W, Xu M, Wang L, et al. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduct Target Ther 2019;4:55. [Crossref] [PubMed]
- Kinoshita M, Ikei N, Shin S, et al. Relationship between human papillomavirus and oncogenes (c-myc, N-myc) amplification in human cervical cancers. Nihon Gan Chiryo Gakkai Shi 1990;25:2828-35. [PubMed]
- Kübler K, Heinenberg S, Rudlowski C, et al. c-myc copy number gain is a powerful prognosticator of disease outcome in cervical dysplasia. Oncotarget 2015;6:825-35. [Crossref] [PubMed]
- Ran S, He X, Jiang Z. Genome-wide association analysis identifying that genes ANXA8 and C10orf11 are candidate genes for sarcopenia. Journal of University of Shanghai for Science and Technology 2020;42:305-10.
- Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 1988;54:5-16. [Crossref] [PubMed]
- Freyd G, Kim SK, Horvitz HR. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 1990;344:876-9. [Crossref] [PubMed]
- Lund K, Petersen JS, Jensen J, et al. Islet expression of Rhombotin and Isl-1 suggests cell type specific exposure of LIM-domain epitopes. Endocrine 1995;3:399-408. [Crossref] [PubMed]
- Dawid IB, Toyama R, Taira M. LIM domain proteins. C R Acad Sci III 1995;318:295-306. [PubMed]
- Michelsen JW, Schmeichel KL, Beckerle MC, et al. The LIM motif defines a specific zinc-binding protein domain. Proc Natl Acad Sci U S A 1993;90:4404-8. [Crossref] [PubMed]
- Xue D, Tu Y, Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science 1993;261:1324-8. [Crossref] [PubMed]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol 2004;5:920-31. [Crossref] [PubMed]
- Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature 1995;374:425-30. [Crossref] [PubMed]
- Porter FD, Drago J, Xu Y, et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 1997;124:2935-44. [Crossref] [PubMed]
- Birk OS, Casiano DE, Wassif CA, et al. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature 2000;403:909-13. [Crossref] [PubMed]
- Liang TS, Zheng YJ, Wang J, et al. MicroRNA-506 inhibits tumor growth and metastasis in nasopharyngeal carcinoma through the inactivation of the Wnt/β-catenin signaling pathway by down-regulating LHX2. J Exp Clin Cancer Res 2019;38:97. [Crossref] [PubMed]
- Zhou F, Gou S, Xiong J, et al. Oncogenicity of LHX2 in pancreatic ductal adenocarcinoma. Molecular Biology Reports 2014;41:8163-7. [Crossref] [PubMed]
- Shi X, Zhan L, Xiao C, et al. miR-1238 inhibits cell proliferation by targeting LHX2 in non-small cell lung cancer. Oncotarget 2015;6:19043-54. [Crossref] [PubMed]
- Song H, Liu J, Wu X, et al. LHX2 promotes malignancy and inhibits autophagy via mTOR in osteosarcoma and is negatively regulated by miR-129-5p. Aging (Albany NY) 2019;11:9794-810. [Crossref] [PubMed]
(English Language Editor: J. Jones)