
Identification of RHEX as a novel biomarker related to progression and immunity of non-small cell lung carcinoma
Introduction
Although the prevalence of lung cancer has been gradually declining over the past decade, it remains one of the most common tumors and the leading cause of cancer-related mortality worldwide (1). Non-small cell lung carcinoma (NSCLC) accounts for the vast majority of lung cancers, of which lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the most common subtypes (2,3). Although the introduction of several molecular targeted drugs and immune checkpoint inhibitors has dramatically changed the therapeutic landscape for NSCLC patients (4,5), some remain refractory to treatment and certain therapeutic drugs are unable to provide long-term remission, resulting in a poor prognosis. Therefore, more specific biomarkers are required for predicting the prognosis and progression of NSCLC.
Concerns regarding the therapeutic response and prognosis of patients with NSCLC are mostly related to immunity (6,7). The tumor microenvironment (TME) is comprised of tumor cells and surroundings such as blood vessels, the extracellular matrix, and other nonmalignant cells such as mesenchymal stem/stromal cells, fibroblasts, pericytes, and immune cells (8). A growing body of studies have closely investigated the impact of immune-related genes and immune cells in the tumor microenvironment (TME) on tumor progression, therapeutics, and clinical outcomes. Baraibar et al. confirmed that differentiation inhibitor 1 (ID1) in KRAS-mutant lung cancer could assist programmed cell death protein 1 (PD-1) inhibitors to impair tumor growth and survival by stimulating programmed cell death protein ligand 1 (PD-L1) expression and increasing tumor-infiltrating CD8+ T cells (9), and several clinical studies have shown that higher levels of intratumoral infiltration of CD4+ and/or CD8+ T cells are associated with longer survival in early NSCLC (10,11). Further, Li et al. demonstrated that regulatory T cells (Tregs) were more efficient than cytokeratin-19 fragments (Cyfra21-1) and carcinoembryonic antigen (CEA) to predict tumor recurrence in patients with NSCLC following radical surgery (12). Therefore, to improve the prognosis of NSCLC patients and to provide reliable information to guide appropriate personalized treatment strategies, it is necessary to identify reliable immunological prognostic or predictive indicators that are closely related to tumor phenotype and immune traits.
In recent years, with the development of genomic technologies, bioinformatics has been widely used to study the molecular mechanisms of diseases and to identify disease-specific biomarkers (13). The advantage of weighted gene co-expression network analysis (WGCNA) is that it can be used to detect the co-expression of highly relevant genes and modules of interest related to clinical traits (14). Differential gene expression analysis identifies differentially expressed genes (DEGs) that may play key roles in human disease between experimental groups and control groups. The single-sample gene set enrichment analysis (ssGSEA) applies the genetic characteristics expressed by immune cell populations to individual cancer samples, and according to the results obtained, the immune infiltration traits of tumors can be extracted and grouped. At present, there are few reports based on the combination of differential gene expression analysis with ssGSEA and WGCNA.
In the present study, based on The Cancer Genome Atlas (TCGA)-NSCLC mRNA expression data, we screened hub genes related to the tumor phenotype and immune traits using a novel approach investigating differential gene expression analysis with ssGSEA and WGCNA. Finally, the regulators of hemoglobinization and erythroid cell expansion (RHEX) were screened as a key gene and related to the prognosis, tumor phenotype, and immunity traits of patients with NSCLC. At present, only one study in solid tumors reported that RHEX expression was higher in lymph node metastasis-positive patients than in lymph node metastasis-negative patients, and it may be a new biomarker for predicting lymph node metastasis in patients with early-stage endometrial cancer (15). Therefore, our study is the first to examine the role of RHEX in NSCLC providing valuable insights for clinicians and researchers globally, and is designed to encourage the further validation of these results to implement their use in clinical practice and ultimately, curb the growing burden of NSCLC. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1316).
Methods
Collection and grouping of NSCLC data
The transcriptome count data and corresponding clinical data of NSCLC were downloaded from the TCGA program (https://portal.gdc.cancer.gov). The data of fragments per kilobase million (FPKM) were first converted from count data and used for immune grouping of NSCLC samples by ssGSEA, which then applied the genetic characteristics expressed by immune cell populations to individual cancer samples. We used the ssGSEA method of R software gene set variation analysis (GSVA) package to analyze the infiltration level of different immune cells, immune-related pathways, and the activity of immune-related functions in the profile data of NSCLC expression. According to the results of ssGSEA, the TCGA-NSCLC samples were divided into a high immune cell infiltration group (Immunity_H group) and low immune cell infiltration group (Immunity_L group) using the R package “hclust”. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Verification of the effectiveness of immune grouping
By Estimation of STromal and Immune cells in MAlignant Tumour tissues using Expression data (ESTIMATE) algorithm, the ESTIMATE score, immune score, stromal score, and tumor purity were analyzed to verify the effect of ssGSEA immune grouping and to draw a clustering heat map and statistical map in the TCGA-NSCLC dataset. Human leukocyte antigen (HLA) and PD-L1 gene expression levels were used to verify differences between the two groups. The CIBERSORT deconvolution algorithm was used to accurately assess the composition of immune infiltrating cells in tumor samples, thereby verifying the differences in the level of immune cell infiltration between the two groups. Finally, to confirm differences in the enrichment pathways between the two groups, we performed GSVA enrichment analysis using “GSVA” R packages. The gene sets of “c2.cp.kegg.v7.1.symbols.gmt” were downloaded from the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) for running GSVA analysis. Statistical significance was set at a P value <0.05.
Identification of DEGs
According to the criteria of |log2FC| ≥1 and adjusted P<0.05, the R package “limma” and “edgeR” were separately utilized to normalize the data and discover DEGs between the Immunity_L and Immunity_H group. Next, the differential analysis was carried out according to the same criteria between the tumor and normal groups. In addition, to make the data types more appropriate for our next analysis, we retained genes with counts per million (CPM) ≥1, then converted the count values into reads per kilobase million (RPKM) values by dividing gene counts by the length of the gene.
WGCNA
We used the “WGCNA” feature in the R package to separately find the module that was most positively related to tumor phenotype or Immunity_H traits and the hub genes in the module. The adjacency matrix was transformed into a topological overlap matrix (TOM), and genes were divided into different gene modules according to the TOM-based dissimilarity measure. Here, we set the soft-thresholding power to 6 (scale free R2=0.90), defined the height as 0.25 and the minimal module size as 50 to identify key modules. Subsequently, the overlapping genes between DEGs and co-expression genes extracted from the co-expression network were used as hub genes and used to identify potential prognostic genes, which are presented as a Venn diagram.
Functional annotation and survival analysis of hub genes
We conducted the Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses using the R package feature “clusterProfiler.” GO terms or KEGG pathways with adjusted P<0.05 were considered significant, and were visualized by the R package feature “GOplot.”
For further screening of key genes, survival analysis was conducted for hub genes using the R package features “survminer” and “survival.” Tumor samples within the TCGA-NSCLC dataset were divided into a high and low expression group, based on the median value of each hub gene to plot the Kaplan-Meier survival curves.
Validation in ArrayExpress database
We downloaded the series matrix files of the dataset (E-MTAB-6043) and the corresponding clinical data of NSCLC from ArrayExpress (http://www.ebi.ac.uk/arrayexpress). As described above, ArrayExpress-NSCLC samples were also divided into high and low immune groups and the grouping was confirmed, as shown in Figure S1. Differential gene expression analysis and Kaplan-Meier survival analysis were then performed in the ArrayExpress-NSCLC dataset.
Immune-related analysis of RHEX expression
We utilized software GSEA 4.1.0 (https://www.gsea-msigdb.org/gsea/downloads.jsp) to perform GSEA analysis. Based on the median RHEX mRNA expression, 1,037 NSCLC samples were divided into high- and low-RHEX expression, and the gene set “h.all.v7.1.symbols” was selected for the enrichment analysis. Permutations were set to 1,000 to obtain a normalized enrichment score, and statistical significance was set at P<0.05.
In addition, CIBERSORT analysis was used to investigate the association between RHEX and TICs. According to the median, 1,037 TCGA-NSCLC samples were first normalized using the voom function and were divided into high and low-RHEX expression groups. LM22 is an annotated gene signature matrix that defines 22 immune cell subtypes and was downloaded from the CIBERSORT web portal (http://cibersort.stanford.edu/), and we ran CIBERSORT locally in the R software. The number of permutations was set to 1,000, and a threshold P value of <0.05 was the criteria for successful computation of a sample. To further confirm the correlation between the expression of RHEX and TICs, we applied the “gene module” of the online tool Tumor IMmune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) which contains 10,897 samples from diverse cancer types available in the TCGA database. Moreover, the correlation between the somatic copy number alteration (SCNA) of RHEX and immune abundance of the leukocytes was explored using the “SCNA module.” The association between RHEX and immune modulators in pan-cancer was depicted through the web portal TISIDB (http://cis.hku.hk/TISIDB/index.php) (16), which is an integrated repository portal for tumor–immune system interactions.
Statistical analysis
Statistical analyses were performed using R software (version 3.6.0, https://www.r-project.org). The survival curves for prognostic analyses were generated using the Kaplan–Meier method, and the log-rank test was used to identify the significance of differences. The correlation between RHEX expression and clinical parameters was analyzed using the R package feature “ggpubr”, and gene expression correlations were analyzed using Spearman’s correlation. For all comparisons, a two-tailed P value <0.05 was considered significant.
Results
Construction and verification of the NSCLC immune groupings
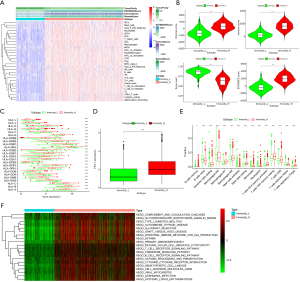
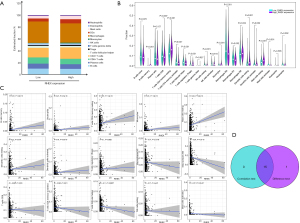
The flowchart of this study is summarized in Figure S2. We obtained 1,037 NSCLC samples and 108 paracancerous samples from the TCGA database, and the ssGSEA method was applied to the NSCLC expression profile data to evaluate the infiltration of immune cells. Using an unsupervised hierarchical clustering algorithm, NSCLC samples were divided into an Immunity_H group (n=758) and Immunity_L group (n=279) (Figure 1A), and to prove the feasibility of this immune grouping strategy, the ESTIMATE algorithm was employed to calculate the ESTIMATE score, stromal score, immune score, and tumor purity. While compared with the Immunity_L group, the Immunity_H group had lower tumor purity, the other scores were higher (Figure 1A) in the latter. The violin diagram also showed a significantly positive correlation between the Immunity_H group and the ESTIMATE score, stromal score, and immune score, and showed a negative correlation with tumor purity (Figure 1B). We also found expression of the HLA family and PD-L1 in the Immunity_H group was significantly higher than in the Immunity_L group (P<0.05) (Figure 1C,1D), and the CIBERSORT method revealed there were more immune cells in the Immunity_H group than in the Immunity_L group (Figure 1E). GSVA enrichment analysis was performed to demonstrate that the above groups differed in the KEGG enrichment pathways. The top 20 enrichment pathways are shown in Figure 1F, and the Immunity_H group was markedly enriched in immune-related pathways. To summarize, the Immunity_H group had higher immune components and lower tumor purity than the Immunity_L group (P<0.05), indicating the NSCLC immune grouping could be used for follow-up analysis.
Analyses of DEGs between the tumor and normal groups and between the Immunity_H and Immunity_L groups
Based on the cutoff criteria of |log2FC| ≥1 and adjusted P<0.05, we analyzed the difference between the tumor group (1,037 cases) and the normal group (108 cases). We found 4,283 DEGs, of which 1,755 and 2,528 were upregulated and downregulated, respectively (Figure 2A). According to the same criteria, 1,277 DEGs were identified in the Immunity_H group compared to that identified in the Immunity_L group, of which 1,077 were upregulated and 200 were downregulated (Figure 2B).
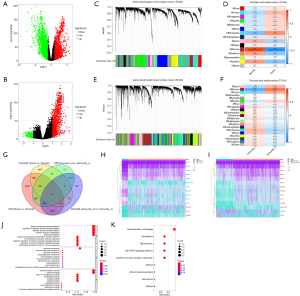
WGCNA and extraction of overlapping genes
Co-expression networks facilitate network-based gene screening methods that can be used to identify hub biomarkers and therapeutic targets. Based on this, to screen co-expression genes most relevant to NSCLC clinical traits (tumor & Immunity_H groups), WGCNA analysis was performed twice to find the key modules most positively related to tumor and immunity traits in TCGA (Figure 2C-2F). From the heatmap of module-trait correlations we identified that the turquoise module was most positively correlated with tumor traits (cor =0.55, P<0.0001), which included 3,232 genes (Figure 2D), and the brown module was the most highly correlated with the Immunity_H trait (cor =0.67, P<0.0001), which included 1,511 genes (Figure 2F).
As shown in Figure 2G, after Venn analysis, 14 co-expressing DEGs were extracted between the DEGs lists and the co-expression modules including IL22RA2, POU6F2, CSF3R, RAB37, RHEX, CX3CR1, FOXI3, ICAM1, CD1E, CD1C, MAP1LC3C, FCER1A, BANK1, and CHI3L2. Finally, we plotted the heat map of differential expression of the 14 hub genes in the tumor versus the normal group (Figure 2H) and in the Immunity_H versus the Immunity_L group (Figure 2I) to show their expression patterns.
Functional enrichment analyses for the 14 DEGs
To gain further insights into the potential functions of the 14 identified DEGs, we conducted GO and KEGG analyses, and a P value <0.05 was considered significant. The top 10 factors of each aspect of GO are shown in Figure 2J. The GO terms were mainly clustered in immune-related pathways, tyrosine phosphorylation, and apoptotic signaling pathways. The significant KEGG pathways are displayed in Figure 2K, including hematopoietic cell lineage, amoebiasis, tight junction, JAK-STAT signaling pathway, asthma, cytokine-cytokine receptor interaction, African trypanosomiasis, ferroptosis, and malaria. Unsurprisingly, these pathways were associated with tumor and immunity traits, which was consistent with our screening approach. These results suggest that these 14 DEGs may be involved in immune-oncology-related functions.
Validation and identification of key genes
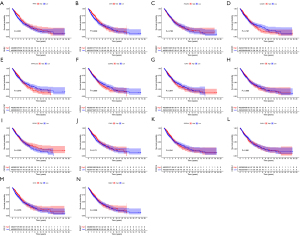
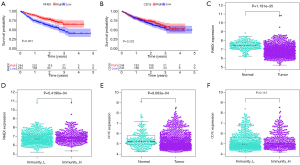
We performed a Kaplan-Meier analysis to determine whether the expression of these genes was related to the overall survival (OS) of patients with NSCLC in TCGA (Figure 3A-3N) and found RHEX (P=0.031) (Figure 3A) and CD1E (P=0.039) (Figure 3B) were associated with a favorable clinical prognosis. Based on the ArrayExpress-NSCLC microarray dataset, we verified the results of the above two genes (Figure 4A,4B) and found that RHEX expression alone was associated with a good prognosis. Simultaneously, we analyzed the expression profiles of RHEX and CD1E between the tumor and the normal group and between the Immunity_H group and Immunity_L group in this dataset (Figure 4C-4F) and again found the expression pattern of RHEX alone was consistent with the TCGA dataset. As only the expression level of RHEX was related to the prognosis, tumor phenotype, and immunity traits of patients with NSCLC in both the databases, we focused on the RHEX gene in subsequent analyses.
Correlation of RHEX expression with clinicopathological parameters in NSCLC patients
The relationship between RHEX expression levels and clinicopathological parameters was explored in TCGA-NSCLC patients (Figure 5A-5F), and the results revealed that RHEX expression was higher in females than in males (Figure 5B). The expression of RHEX in early-stage NSCLC cases (stage I) was significantly higher than that in the middle- and advanced-stage cases (Figure 5C) and was significantly higher in the T1 stage than that in other T stages (Figure 5D). However, the correlation between other parameters and RHEX expression was not significant. Overall, these results suggest that RHEX expression levels decline as NSCLC progresses.
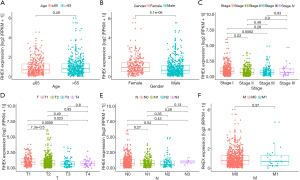
RHEX is a potential immune-predictive indicator
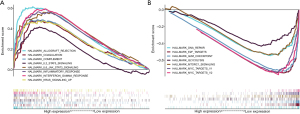
To further confirm the functions of RHEX we performed a GSEA by separating patients with NSCLC into two groups according to the median RHEX expression. As presented in Figure 6A and Table S1, in the high-RHEX expression phenotype, eight gene sets were upregulated and significantly enriched in immune-related activities, while in the low-RHEX expression group, the genes were mainly enriched in cell cycle-related pathways (Figure 6B and Table S1). These results suggest that as RHEX expression decreases, the status of the TME shifts from being immune-predominant to cell cycle control-dominant, which indicates RHEX might be a potential indicator of immune status.
Significant correlation between RHEX and TICs in the TME
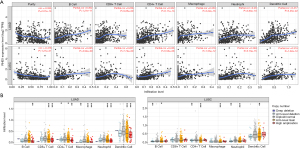
To confirm the correlation between RHEX expression and the immune microenvironment, we conducted a CIBERSORT analysis and referred to the TIMER. The CIBERSORT algorithm applied to the 22 immune cell subtypes helped assess their differing concentrations in the high- and low-RHEX expression groups. As the estimated proportion of CD4+ naive T cells was not found in the eligible sample group, it was not included in the subsequent analysis. Figure 7A shows the proportion of 14 subpopulations of immune cells in the high- and low-RHEX expression groups and reveals the proportion of TICs in the two groups differs. Moreover, differential analysis using the Wilcoxon test indicated that RHEX expression was correlated with multiple TICs as shown in Figure 7B. Spearman’s correlation analysis found a significant correlation between RHEX and 15 TICs (Figure 7C) and the results from the differential and correlation analyses showed that the 15 types of TICs were correlated with the expression of RHEX (Figure 7D). Among them, eight types were positively correlated with RHEX expression, including memory B cells, CD4+ memory resting T cells, Tregs, monocytes, resting/activated dendritic cells, neutrophils, and resting mast cells, while seven TICs were negatively correlated with RHEX expression, including activated CD8+ T cells, follicular helper T cells, CD4+ memory activated T cells, macrophage M0/M1, resting NK cells, and activated mast cells. Taken together, these results strongly suggest RHEX is important for immune cell infiltration in tumor pathology.
To confirm the above results, the TIMER tool was used for analysis. As a result, the expression level of RHEX was negatively correlated with tumor purity, indicating their comparative enrichment outside tumor cells may be attributable to the enrichment patterns of RHEX in the TME, while RHEX was positively correlated with the infiltration levels of six immune cells both in LUAD and LUSC (Figure 8A). In addition, different mutational forms of RHEX were associated with immune infiltration of these six TICs (Figure 8B), which also reveals its influence on the NSCLC immune microenvironment. In particular, arm-level gain of RHEX was associated with substantially lower levels of six immune infiltrates than normal RHEX expression in NSCLC.
Relationship between RHEX expression and immune modulators in NSCLC
To investigate the potential impact of RHEX expression on NSCLC immunotherapy, we systematically analyzed correlations between RHEX expression and three types of immune modulators described in a previous study conducted by Charoentong et al. through the TISIDB database (17). Intriguingly, we found RHEX expression was positively correlated with most of the immune modulators, including immunoinhibitors, immunostimulators, and major histocompatibility complex (MHC) molecules in pan-cancer (Figure 9A-9C). More importantly, we observed that RHEX was positively correlated with almost all MHC molecules in NSCLC, and very few immune modulators showed negative correlation with RHEX expression in NSCLC. These results indicate that RHEX might play a pivotal role in co-regulating the NSCLC TME.
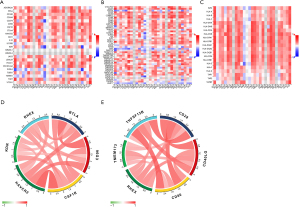
To further explore and confirm the relationships between RHEX and immune checkpoint members (immunoinhibitor and immunostimulator), correlation analyses were performed in the TCGA database (Figure 9D,9E; Table S2) and, as expected, RHEX was positively correlated with most of the checkpoint members (Table S2). The top five immunoinhibitors and immunostimulators most associated with RHEX expression are respectively plotted in the circle diagrams (Figure 9D,9E). Thus, our findings revealed that RHEX might manipulate anti-tumor immune responses through co-regulation with RHEX-related immune checkpoint molecules, thereby lending support to the use of combination cancer immunotherapy targeting these molecules in future studies.
Discussion
Although various therapeutic strategies such as surgery, chemotherapy, radiotherapy, and immunotherapy have been used to improve the clinical efficacy of NSCLC patients for many years, the prognosis remains less than satisfactory, prompting us to search for the underlying molecular mechanisms related to this condition. Multiple investigators have reported that immune-related genes can be used as prognostic indicators for NSCLC (18-21). A growing body of evidence shows the importance of biomarkers, especially genes, in determining the outcomes of cancer, offering new opportunities for integrating this information into therapeutic algorithms, especially those related to immunity.
Our study is the first to investigate differential gene expression analysis combined with WGCNA and ssGSEA to explore novel tumor- and immune-related genes. We successfully identified 14 overlapping genes that were significantly related to the tumor phenotype and immunity, among which, RHEX alone had the same expression trend and prognostic value in both the TCGA and ArrayExpress databases. Compared with the tumor and Immunity_L groups, RHEX mRNA expression was significantly higher in the normal group and in the Immunity_H group. In addition, this is the first study to report a consistent association between increasing RHEX mRNA levels and favorable clinical prognosis in NSCLC patients.
RHEX (also called C1orf186) was originally identified as a transduction factor of the erythropoietin-erythropoietin receptor (EPO-EPOR) signaling pathway, modulating human erythroid progenitor cell (hEPCs) expansion and promoting erythroid cell differentiation (22). RHEX is well-conserved in humans and other primates but not in the genomes of rats, mice, or lower vertebrates. RHEX mRNA is also expressed in multiple primary human tissues, including the brain, liver, lung, and peripheral blood cells (monocytes, T cells, neutrophils, and platelets), and is highly expressed in primary hEPCs and the kidney (15,22,23). RHEX is a plasma membrane protein, whose predicted domains include an amino-terminal (NT) hydrophobic region and two carboxy-terminal candidate growth factor receptor-bound protein 2 (GRB2)-binding sites (22,24). RHEX protein proved to be upregulated by EPO ≥20-fold in its phosphorylation at phosphotyrosine (PY) 132 and PY141 sites, which were also implicated in binding GRB2. Beyond this, RHEX protein has been shown to comprise a likely Janus kinases-2 (JAK2) PY target and to co-immunoprecipitate with EPOR and JAK2, both of which indicate it may be involved in crosstalk between the EPOR/JAK signaling pathways (22,25). EPO/EPOR ligation is known to activate JAK2 kinase, JAK2 phosphorylation of EPOR cytoplasmic PY motifs, and canonical STAT, PI3K, and RAS/MEK/ERK signal transduction pathways (26,27). JAK2 plays an important role in the promotion of multiple oncogenic phenotypes encompassing tumorigenesis, proliferation, invasion, metastasis, and immune evasion (28). Therefore, based on the above literature mining, we can speculate that RHEX may affect multiple immune tumor-related signaling pathways through the EPOR/JAK signaling pathways.
Presently, few studies have been conducted on RHEX in solid tumors, and its role in NSCLC has not yet been elucidated. Given that our knowledge on the role of RHEX in cancer is limited, herein we aimed to dissect its biological functions in NSCLC and reveal its associated regulatory pathways by performing a comprehensive analysis of open-access databases. Co-expressed genes act synergistically in biological processes under strict regulatory control and have an advantage during adaptive evolution (29). Based on the functional enrichment analyses of the 13 genes in the RHEX co-expression module, we attempted to elucidate the biological roles of RHEX. First, we identified some functional terms that corresponded to previous studies on RHEX, especially the tyrosine phosphorylation pathway, hematopoietic cell lineage, and JAK-STAT signaling pathway, which confirmed the reliability of our analytical method. Second, functional enrichment analysis revealed that RHEX was correlated with a variety of immune response pathways, including T cell activation, lymphocyte activation, antigen processing and presentation, leukocyte migration, and cytokine and cytokine receptor interaction, which indicated it might promote TME immune infiltration by these functional terms. Third, following enrichment analysis, we finally found programmed cell death-related terms, such as extrinsic apoptosis signaling pathway and ferroptosis, which suggested RHEX might be involved in anti-tumor mechanisms. These results strongly indicated RHEX may have complex regulatory roles in NSCLC tumors.
To further explore the biological functions of RHEX in NSCLC, we conducted a GSEA and found the samples with high-RHEX expression were positively correlated with signaling pathways implicated in various immune-oncology-related pathways, especially the IL-6/STAT3 pathway. We speculated that RHEX may participate in anti-tumor immune mechanisms by interfering with abnormally activated JAK-related signaling pathways in NSCLC. Previous studies have shown that abnormal activation of the IL-6/STAT3 pathway could induce the expression of PD-L1, enhance tumor cell proliferation, and inhibit tumor cell apoptosis in NSCLC (30,31). In addition, Cai et al. reported that decreased expression of JAK1 was associated with immune infiltration and a poor prognosis in LUAD tissues (32). While the specific regulatory mechanism of RHEX involved in JAK-related signaling pathways needs to be verified through more detailed experiments, the low expression group was mainly enriched in multiple gene sets with well-established roles in NSCLC development, including G2M_CHECKPOINT, MYC_TARGETS_V1/V2, E2F_TARGETS, DNA_REPAIR, MTORC1_SIGNALING, and GLYCOLYSIS. It is widely believed that abnormal cell proliferation is a hallmark of malignant tumors, and tumor cells often show changes in the expression of genes that directly regulate their cell cycles (33). Each replication of DNA in cancer cell cycles may cause a large amount of damage, including DNA substitutions or deletions (34). Therefore, the mechanisms of DNA repair and cell cycle examinations are particularly important. Notably, as a proto-oncogene, MYC encodes a transcription factor that regulates gene networks controlling cell cycle progression, apoptosis, and cellular transformation (35,36), and previous studies have indicated that it is able to promote the proliferation, metastasis, and metabolism of NSCLC by regulating multiple pathways (37-40). In addition, the roles of E2F_TARGETS, MTORC1_SIGNALING, and GLYCOLYSIS in NSCLC have been reported in several studies (41-43). Above all, we infer that high-RHEX expression can participate in immunomodulation and promote the prognosis of patients, while a decrease in RHEX expression is beneficial for tumor progression and leads to a poor prognosis. In addition, we verified that RHEX mRNA levels were inversely correlated with the T stage status of NSCLC patients. Thus, our data confirmed the downregulation of RHEX coincides with the advancing stage of NSCLC and the conversion of TME from an immune-predominant to a cell cycle control-dominant status and supported the proposition that RHEX might play an important anti-tumor role in NSCLC tumors.
With the development of scientific research, immune cell infiltration in the TME could help elucidate the mechanisms behind tumor development. Here, we could only determine the significant correlations between immune cell infiltration and RHEX expression in tumors, while the relationship between cause and effect was difficult to establish. Overexpression of RHEX mRNA leads to bidirectional changes in TIC infiltration levels, namely, the immune abundance of memory B cells, CD4+ memory resting T cells, Tregs, monocytes, resting/activated dendritic cells, neutrophils, and resting mast cells, which is consistent with the results of the TIMER database. Conversely, CD8+ T cells, follicular helper T cells, CD4+ memory activated T cells, macrophage M0/M1, resting NK cells, and activated mast cells were observed to decrease. The immune infiltration levels of antigen-presenting cells such as macrophages, monocytes, dendritic cells, and B cells were significantly correlated with the RHEX expression level, which was consistent with the previously mentioned role of RHEX in promoting immune infiltration in the TME by enhancing antigen processing and presentation and lymphocyte activation. However, we also found contradictory results in the correlation analysis of RHEX expression levels and abundance of CD8+ T cell infiltration, which we speculated might be due to the heterogeneity of data processing, while the specific mechanism remains to be further confirmed by experiments. In addition, we also found a strong association between RHEX copy number variation and immune infiltrates. The finding further revealed a correlation between RHEX expression and immune infiltration, suggesting a potential mechanism by which RHEX alterations predict the response to immunotherapy. These results demonstrated the influence of RHEX on immune processes in NSCLC is a comprehensive biological effect. As mentioned earlier (10-12,44), some studies have confirmed that the occurrence and progression of tumors can be regulated by TICs in the TME, which are promising targets for treatment and the basis of immunotherapy. Therefore, the crosstalk between RHEX and TICs in NSCLC suggests that RHEX could be used as a potential biomarker for therapeutic strategy design and immune state prediction for NSCLC patients in the future.
Finally, we estimated the association of RHEX with immune checkpoint members to further explore its synergistic role in NSCLC-induced immune responses. Interestingly, our results revealed that immunomodulators, including stimulator and suppressor molecules, were positively related to RHEX expression simultaneously in NSCLC, which is a good reflection of the complexity of the TME. Studies have shown that the complex interplay between cancer and its TME is also responsible for resistance to immunotherapy (45). Hence, the complex interaction interplay between RHEX expression, immune inhibitors, and immune stimulators in the TME might be one possible explanation for NSCLC immunotherapy resistance. In conclusion, our findings revealed that RHEX might manipulate anti-tumor immune responses through co-regulation with RHEX-related immune checkpoint molecules, and suggests immunotherapy, by both blocking inhibitory checkpoints and activating stimulatory pathways, should be strongly considered.
Notably, the main limitation of our study is that it was based on preliminary data generating predictions. Although we provided a comprehensive bioinformatics analysis to identify potential key genes, RHEX expression needs to be validated between normal and tumor and between high and low levels of immune cell infiltration groups. Furthermore, the specific mechanisms by which RHEX regulates immune cell infiltration and the anti-tumor response requires further investigation through a series of experiments.
Conclusions
In this study, we found that RHEX significantly correlated with the prognosis, immune infiltration, and immunomodulators. Our findings shed light on the important role of RHEX in NSCLCs and provided insight into the potential relationship and underlying mechanism between RHEX and tumor-immune system interactions.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1316
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1316). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet 2011;378:1727-40. [Crossref] [PubMed]
- Hu Z, Hou D, Wang X, et al. TSPAN12 is overexpressed in NSCLC via p53 inhibition and promotes NSCLC cell growth in vitro and in vivo. Onco Targets Ther 2018;11:1095-103. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Correction to: "Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up Ann Oncol 2019;30:863-70. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- Wood SL, Pernemalm M, Crosbie PA, et al. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev 2014;40:558-66. [Crossref] [PubMed]
- Meng W, Xue S, Chen Y. The role of CXCL12 in tumor microenvironment. Gene 2018;641:105-10. [Crossref] [PubMed]
- Baraibar I, Roman M, Rodríguez-Remírez M, et al. Id1 and PD-1 Combined Blockade Impairs Tumor Growth and Survival of KRAS-mutant Lung Cancer by Stimulating PD-L1 Expression and Tumor Infiltrating CD8+ T Cells. Cancers (Basel) 2020;12:3169. [Crossref] [PubMed]
- Hiraoka K, Miyamoto M, Cho Y, et al. Concurrent infiltration by CD8+ T cells and CD4+ T cells is a favourable prognostic factor in non-small-cell lung carcinoma. Br J Cancer 2006;94:275-80. [Crossref] [PubMed]
- Zhuang X, Xia X, Wang C, et al. A high number of CD8+ T cells infiltrated in NSCLC tissues is associated with a favorable prognosis. Appl Immunohistochem Mol Morphol 2010;18:24-8. [Crossref] [PubMed]
- Li SJ, Wu YX, Chen HL, et al. Correlation of CD4+CD29+ regulatory T cells with recurrence and survival time in patients with non-small cell lung cancer. Nan Fang Yi Ke Da Xue Xue Bao 2016;36:1215-20. [PubMed]
- Can T. Introduction to bioinformatics. Methods Mol Biol 2014;1107:51-71. [Crossref] [PubMed]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17.
- Huang CY, Liao KW, Chou CH, et al. Pilot Study to Establish a Novel Five-Gene Biomarker Panel for Predicting Lymph Node Metastasis in Patients With Early Stage Endometrial Cancer. Front Oncol 2020;9:1508. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Schmidt LH, Kümmel A, Görlich D, et al. PD-1 and PD-L1 Expression in NSCLC Indicate a Favorable Prognosis in Defined Subgroups. PLoS One 2015;10:e0136023. [Crossref] [PubMed]
- Li R, Liu X, Zhou XJ, et al. Identification and validation of the prognostic value of immune-related genes in non-small cell lung cancer. Am J Transl Res 2020;12:5844-65. [PubMed]
- Wang Q, Gu J, Wang L, et al. Genetic associations of T cell cancer immune response-related genes with T cell phenotypes and clinical outcomes of early-stage lung cancer. J Immunother Cancer 2020;8:e000336. [Crossref] [PubMed]
- Massarelli E, Lam VK, Parra ER, et al. High OX-40 expression in the tumor immune infiltrate is a favorable prognostic factor of overall survival in non-small cell lung cancer. J Immunother Cancer 2019;7:351. [Crossref] [PubMed]
- Verma R, Su S, McCrann DJ, et al. RHEX, a novel regulator of human erythroid progenitor cell expansion and erythroblast development. J Exp Med 2014;211:1715-22. [Crossref] [PubMed]
- Broxmeyer HE. RHEX in the mix of erythropoietin signaling molecules. J Exp Med 2014;211:1704. [Crossref] [PubMed]
- Neumann K, Oellerich T, Urlaub H, et al. The B-lymphoid Grb2 interaction code. Immunol Rev 2009;232:135-49. [Crossref] [PubMed]
- Held MA, Greenfest-Allen E, Su S, et al. Phospho-PTM proteomic discovery of novel EPO- modulated kinases and phosphatases, including PTPN18 as a positive regulator of EPOR/JAK2 Signaling. Cell Signal 2020;69:109554. [Crossref] [PubMed]
- Watowich SS. The erythropoietin receptor: molecular structure and hematopoietic signaling pathways. J Investig Med 2011;59:1067-72. [Crossref] [PubMed]
- Wojchowski DM, Sathyanarayana P, Dev A. Erythropoietin receptor response circuits. Curr Opin Hematol 2010;17:169-76. [PubMed]
- Balko JM, Schwarz LJ, Luo N, et al. Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci Transl Med 2016;8:334ra53. [Crossref] [PubMed]
- Niehrs C, Pollet N. Synexpression groups in eukaryotes. Nature 1999;402:483-7. [Crossref] [PubMed]
- Zhang N, Zeng Y, Du W, et al. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int J Oncol 2016;49:1360-8. [Crossref] [PubMed]
- Zhang X, Zeng Y, Qu Q, et al. PD-L1 induced by IFN-γ from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer. Int J Clin Oncol 2017;22:1026-33. [Crossref] [PubMed]
- Cai J, Deng H, Luo L, et al. Decreased expression of JAK1 associated with immune infiltration and poor prognosis in lung adenocarcinoma. Aging (Albany NY) 2020;13:2073-88. [Crossref] [PubMed]
- Sherr CJ. Cancer cell cycles. Science 1996;274:1672-7. [Crossref] [PubMed]
- Barnes JL, Zubair M, John K, et al. Carcinogens and DNA damage. Biochem Soc Trans 2018;46:1213-24. [Crossref] [PubMed]
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature 2001;411:342-8. [Crossref] [PubMed]
- Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta 2015;1849:506-16. [Crossref] [PubMed]
- Chen S, Gu T, Lu Z, et al. Roles of MYC-targeting long non-coding RNA MINCR in cell cycle regulation and apoptosis in non-small cell lung Cancer. Respir Res 2019;20:202. [Crossref] [PubMed]
- Rapp UR, Korn C, Ceteci F, et al. MYC is a metastasis gene for non-small-cell lung cancer. PLoS One 2009;4:e6029. [Crossref] [PubMed]
- Zhang E, Li W, Yin D, et al. c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer. Tumour Biol 2016;37:4007-15. [Crossref] [PubMed]
- Liu L, Lei B, Wang L, et al. Protein kinase C-iota-mediated glycolysis promotes non-small-cell lung cancer progression. Onco Targets Ther 2019;12:5835-48. [Crossref] [PubMed]
- Xu Y, Huang Y, Weng L, et al. Effects of single-nucleotide polymorphisms in the mTORC1 pathway on the risk of brain metastasis in patients with non-small cell lung cancer. J Cancer Res Clin Oncol 2020;146:273-85. [Crossref] [PubMed]
- Zeng C, Wu Q, Wang J, et al. NOX4 supports glycolysis and promotes glutamine metabolism in non-small cell lung cancer cells. Free Radic Biol Med 2016;101:236-48. [Crossref] [PubMed]
- Wang H, Wang L, Zhang S, et al. Downregulation of LINC00665 confers decreased cell proliferation and invasion via the miR-138-5p/E2F3 signaling pathway in NSCLC. Biomed Pharmacother 2020;127:110214. [Crossref] [PubMed]
- Jackute J, Zemaitis M, Pranys D, et al. Distribution of M1 and M2 macrophages in tumor islets and stroma in relation to prognosis of non-small cell lung cancer. BMC Immunol 2018;19:3. [Crossref] [PubMed]
- Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res 2020;30:507-19. [Crossref] [PubMed]
(English Language Editor: B. Draper)


