
Focal pyroptosis-related genes AIM2 and ZBP1 are prognostic markers for triple-negative breast cancer with brain metastases
Introduction
According to the latest data on the global cancer burden, breast cancer has surpassed lung cancer as the most common cancer in the world, and its mortality rate ranks first among female malignancies (1). Triple-negative breast cancer (TNBC) refers to estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2) negative subtypes of breast cancer, and the incidence of invasive breast cancer is about 12–17% (2). Studies have shown that compared with other subtypes of breast cancer, TNBC has a higher risk of recurrence after surgery and is prone to brain metastasis (BM) (3). When TNBC patients are diagnosed, they often have high tumor grades and are in the late stages of the disease. Due to the lack of ER, PR, and HER-2 receptors in TNBC patients, conventional endocrine therapy and targeted therapy are not very effective, and there is a lack of effective treatment methods. TNBC patients have worse disease-free survival and overall survival compared with luminal and HER-2 positive subtype breast cancers (4). The mechanism of brain metastases of TNBC has not been clear, but there are several ways be accepted by the researchers. For example, grant breast circulating tumor cells (CTCs) extravasation into de brain parenchyma (5), organ-specific colonization dependent on the interaction between cancer cells and their microenvironment (6) or undergo genetic and epigenetic changes to gain brain metastatic competence and so on (7). As the number of TNBC patients increases, so does the number of patients with BM. Thus, the challenging search for effective therapeutic targets and prognostic biomarkers has become the focus of TNBC research.
Pyroptosis is a type of programmed cell death characterized by an innate immune response that relies on caspase and classical inflammasome (8). Studies have shown that pyroptosis can inhibit the occurrence and development of tumors, promote inflammatory death, and form a microenvironment suitable for the growth of tumor cells that promotes tumor growth (9). Several studies have pointed out that apoptosis-related molecules are closely related to the growth, invasion, and metastasis of breast cancer. Storr et al. found that the expression level of Gasdermin B (GSDMB) in breast cancer cells was higher than that in normal breast tissue, and was associated with a high metastasis rate and a low survival rate in patients. Further, cell experiments have shown that interleukin 1 beta (IL-1β) produced by macrophages promotes the migration and lymphatic adhesion of breast cancer cells (10). Notably, the activation process of pyrogen death involves the release of a large number of inflammatory mediators, such as IL-1β and IL-18, which results in a cascade of inflammatory responses that affect the tumor microenvironment (TME). In addition, the low expression of Gasdermin E (GSDME) in breast cancer cells is associated with a significantly increased risk of lymph node metastasis (11). Thus, the expression level of GSDME may induce the pyroptosis of tumor cells. Immunology-related studies have shown that human umbilical cord mesenchymal stem cells can induce cell apoptosis and Michigan Cancer Foundation-7 cell death by secreting IL-1β (12). In terms of immunity, Homo sapiens antiviral innate immune response receptor (RIG-I) mediated innate immune response leads to breast cancer cell death and is associated with the increased expression of inflammatory cytokines induced by pyroptosis (13). These studies suggest that cell pyroptosis plays an important role in the occurrence, development, and prognosis of breast cancer, and may be involved in the regulation of the TME.
Changes in the TME are widely considered an important target that can affect the prognosis of TNBC patients. The TME is composed of cellular and non-cellular compartments. Cancer cells, immune cells, blood vessels, and lymphatics, and fibroblasts comprise the cellular portion of the TME. The non-cellular portion encompasses cytokines, chemokines, mediators, and growth factors. The TME is necessary for in-vasion, metastasis, and settling in a distant location mediates the process by which tumors involve the central nervous system (14). Due to the high-infiltration levels of immune cells in the TNBC tumor microenvironment, immunotherapy has quickly become an emerging treatment method for TNBC patients. However, we found that there is currently a lack of research on the TME and pyroptosis genes of TNBC BM. In this study, bioinformatics method was used to evaluate the expression and prognostic role of pyroptosis genes in BM TNBC, and to explore the mechanism of focal death related to brain metastasis in TNBC patients. Additionally, the infiltration of immune cells in the TME was analyzed to provide a prediction and therapeutic target for the clinical treatment of TNBC BM. We present the following article in accordance with the STREGA reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-2182).
Methods
Datasets
Ribonucleic acid-sequencing (RNA-seq) transcriptome data and the related clinical data of patients with primary TNBC and TNBC BM were obtained from the Gene Expression Omnibus (GEO) database, and standardization was performed on RNA-seq gene expression data for the subsequent analysis. The GSE12276 data set (which comprised 169 patients with TNBC without BM and 35 patients with TNBC with BM, and included their survival data) and the GSE52604 data set (which comprised 20 patients with TNBC without BM and 35 patients with TNBC with BM, but no survival data) were included. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differential expressed genes screening based on pyroptosis genes
First, the GSE12276 and GSE52604 data sets were analyzed for differential genes with or without BM. Pyroptosis genes are derived from the Gene Set Enrichment Analysis (GSEA) database. And then, the intersection genes of DEGs in GSE12276 data set, DEGs in GSE52604 data set and pyroptosis-related genes were seem as TNBC BM related pyroptosis genes.
Survival analysis
The expression and survival of the DEGs in the tumors were analyzed to evaluate the prognostic effect of DEGs. The Kaplan-Meier method was used to plot survival curves. A log-rank P value <0.05 was considered statistically significant.
Immune infiltration and correlation analysis
The single sample gene set enrichment analysis (ssGSEA) algorithm was used to calculate the level of immune infiltration of primary TNBC and TBNC BM. Additionally, the correlations between DEGs and immune cells were analyzed. Finally, the content of immune cells and the prognosis of patients were evaluated.
Drug sensitivity analysis
The TNBC gene expression matrix of the GEO database was used to calculate the sensitivity of the samples to drugs based on the gene expression amount of the samples, and the sensitivity of the drugs was determined using the rank-sum test.
Statistical analysis
All data were analyzed using the R language software package (https://www.r-project.org/). Wilcoxon rank and inspection comparisons of primary TNBC and TNBC BM were conducted to determine differences in gene expression using the Kaplan-Meier survival curve drawing method. Genetic variations with the logarithmic ratio change of |logFC| >0.3 were considered significant. A P value <0.05 was considered statistically significant.
Results
Differential gene screening based on pyroptosis
We first performed a differential gene analysis on the genes of TNBC and TNBC BM in the GSE12276 and GSE52604 data sets, and found that 456 genes were differentially expressed. To further understand the relationship between the pyroptosis genes and the 456 different genes, we conducted an intersection analysis, and found that in DEGs, absent in melanoma 2 (AIM2) and Z-deoxyribonucleic acid-binding protein 1 (ZBP1) were pyroptosis genes (see Figure 1). We hypothesized that these 2 pyroptosis genes might be related to the metastasis and prognosis of TNBC, and thus conducted further investigations.
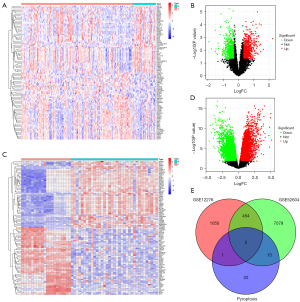
Survival analysis
To understand the prognostic value of AIM2 and ZBP1 in TNBC primary tumor and TNBC BMs, we first performed an expression analysis. The results showed that AIM2 and ZBP1 were significantly overexpressed in TNBC BM in the GSE12276 and GSE52604 data sets (see Figure 2A-2D). A survival analysis showed that patients with high ZBP1 expression had a better prognosis than low expression patients (P=0.036), and patients with high AIM2 expression had a worse prognosis (P=0.075; see Figure 2E,2F). This suggests that AIM2 and ZBP1 play important roles in the occurrence, development and prognosis of TNBC, but the roles of AIM2 and ZBP1 may be opposite to one another.

Immune infiltration and correlation analysis
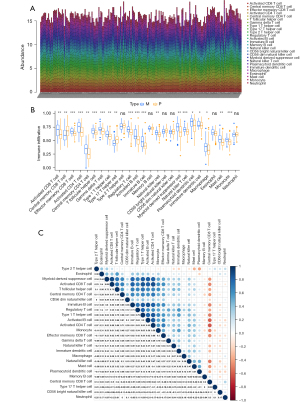
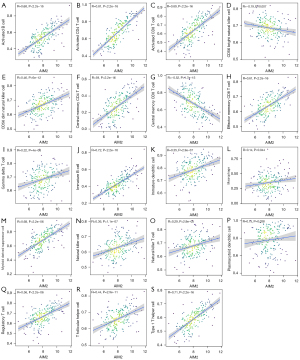
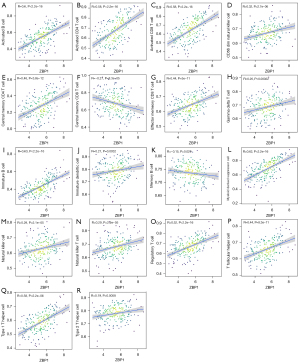
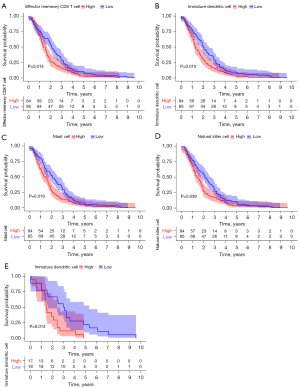
The ssGSEA algorithm was used to evaluate the level of immune infiltration in TNBC primary tumors and TNBC BMs. The results showed that TNBC had a rich infiltration of immune cells, including activated cluster of differentiation 8 (CD8) T cells, effector memory CD8 T cells, activated CD4 T cells, central memory CD4 T cells, T follicular helper cells, gamma delta T cells, type 1 T helper cells, type 2 T helper cells, regulatory T cells, activated B cells, immature B cells, natural killer cells, myeloid-derived suppressor cells, natural killer T cells, plasmacytoid dendritic cells, Immature B cell, and Immature dendritic cell. The infiltration levels of dendritic cells, macrophages, mast cells, and monocytes in primary TNBC tumors were significantly lower than those in TNBC BMs. However, the expressions of central memory CD8 T cells and type 17 T helper cells in primary TNBC tumor was significantly higher than those in TNBC BMs. Second, TNBC was positively correlated with the level of most immune cell infiltrates, monocytes, effector memory CD8 T cells, gamma delta T cells, natural killer T cells, immature dendritic cells, and natural killer cells (see Figure 3). The correlation analysis between AIM2 and ZBP1 and immune cells showed that AIM2 and ZBP1 were significantly correlated with the levels of most immune cells (see Figures 4,5). The analysis of immune cell survival showed that high infiltrating of effector memory CD8 T cells (P=0.014), Immature dendritic cells (P=0.010), mast cells (P=0.019), and natural killer cells (P=0.039) were lead to poor prognosis in TNBC patients without BM. Moreover, high infiltrating of immature dendritic cells (P=0.014) also lead to poor prognosis in TNBC BM patients (see Figure 6).
Drug sensitivity analysis
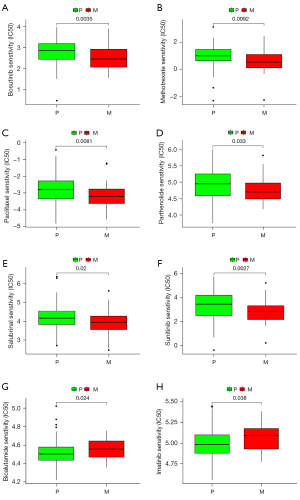
The sensitivity of the drugs in primary TNBC and TNBC BM was analyzed, and the data sets were analyzed. The results showed that Bosutinib (P=0.0035), Methotrexate (P=0.0092), Paclitaxel (P=0.0081), Parthenolide (P=0.033), Salubrinal (P=0.02), and Sunitinib (P=0.0027) were more sensitive to primary TNBC, while Bicalutamide (P=0.024) and Imatinib (P=0.038) were more sensitive to TNBC BM (see Figure 7). These findings have significant implications for treatment decisions for patients with TNBC.
Discussion
Cranial computer tomography, magnetic resonance imaging, positron emission computed tomography and lumbar puncture examination were the diagnostic methods of breast cancer patients with brain metastasis. Compared with computer tomography and positron emission computed tomography, magnetic resonance imaging is more effective in the diagnosis of small intracranial lesions. Nevertheless, these methods are all traditional diagnostic methods. At present, we urgently need new biomarkers to diagnose patients with brain metastasis. In this study, we conducted a bioinformatics analysis of TNBC and TNBC BM, and identified AIM2 and ZBP1 as important genes related to the death of TNBC BM patients. There were significant expression differences of AIM2 and ZBP1 in primary lesions and BMs. Patients with a high expression of AIM2 had a worse prognosis than low expression, while patients with a high expression of ZBP1 had a better prognosis than low expression. These results suggest that AIM2 and ZBP1 may be effective biomarkers in patients with TNBC BM.
AIM2, which is a member of the IFI20x /IFI16 family, plays a putative role in tumorigenic reversion and may control cell proliferation. AIM2 is a cytoplasmic innate immune receptor that assembles inflammasome complexes corresponding to cytoplasmic double-stranded deoxyribonucleic acid (DNA), driving the secretion of IL-1β and IL-18, which in turn induces cell pyroptosis (15). In 2 independent studies of colorectal cancer, AIM2 was found to significantly inhibit colon stem cell proliferation and promote cell death by inhibiting the phosphatidylinositol-3-kinase and protein kinase B (PI3K/Akt) pathway, and is believed to play a regulatory role primarily through the NOD-like receptor family CARD domain containing 3 (NLRC3) (16,17).
AIM2 delays the development of hepatocellular carcinoma. The silencing or overexpression of AIM2 in hepatocellular carcinoma was found to have an anti-cancer effect by forming inflammatory corpuscles and inducing cell pyroptosis, mainly by inhibiting the tumorigenicity of immune-impaired nude mice by inhibiting the mechanistic target of rapamycin (mTOR-S6K1) pathway (18,19). In addition, the disruption of mitochondrial iron metabolism in the absence of phosphatase and tensin homolog induced kinase 1 (PINK1) and parkin ubiquitin protein ligase 2 (PARK2) induces the activation of AIM2 inflammasomes in pancreatic ductal carcinoma and promotes the upregulation of programmed cell death ligand 1 (PD-L1) (20). These studies suggest that AIM2 plays an important role in tumorigenesis and disease progression. Studies of breast cancer have shown that AIM2 inhibits human breast cancer cell proliferation in vitro and breast tumor growth in mouse models, and that AIM2 is associated with tumor suppressor activity (21). Currently, very few studies have explored the relationship between AIM2 and TNBC, and its function in tumors has not yet been fully elucidated.
ZBP1 encodes Z-DNA binding protein, which plays a role in the innate immune response by binding with exogenous DNA and inducing the production of type I interferon. Once ZPB1 is activated by binding with Z-RNA, ZBP1 interacts with RIPK3 to induce cell pyroptosis (22). ZBP1 family proteins are usually silenced or suppressed in normal adult tissues, but studies have shown that the reactivation or increased expression of this gene can be detected in breast cancer, colon cancer, and non-small cell lung cancer, and it is considered to be a characteristic marker of these cancers (23-25). Subsequent studies have shown that ZBP1 activation regulates beta-actin messenger RNA (mRNA) localization, maintains cell polarity and directional movement, and inhibits the chemotaxis and metastasis of breast cancer cells (26-28). In-vitro experiments have demonstrated that blocking the binding of β-catenin to the ZBP1 promoter inhibits the expression of ZBP1, leading to the increased proliferation and migration of metastatic breast cancer cells (29). ZBP1 is essentially a major mRNA regulator that plays an important role in maintaining the stability of non-metastatic phenotypes (30). All these findings suggest that ZBP1 plays an important role in the metastatic behavior of breast cancer cells; however, its role in TNBC had not previously been examined.
Tumor cells and their microenvironment are a functional together. Tumor cells and the TME interact and influence the occurrence and development of tumors. With the development of TME research, researchers have shown that immune cells in the TME play a complex and important role in tumor progression. In this study, AIM2 and ZBP1 were identified as important genes related to TNBC BM. The results showed that AIM2 was positively correlated with tumor-associated macrophages and most T cell types, while AIM2 was negatively correlated with CD56 bright natural killer cells and central memory CD8 T cells. The relationship between expressed level of ZBP1 and infiltration level of immune cells was similar to that of the AIM2, the difference is that there was a negative correlation between ZBP1 and the infiltration level of central memory CD8 T cells and memory B cells. Among numerous immune cells, tumor-associated macrophages account for more than 50% of solid tumor components and participate in the whole process from the occurrence to the metastasis of TNBC, and thus have potential value in the prognosis of TNBC (31-33). In the TME, the abundant invasive level of tumor-associated macrophages suggests a higher risk of tumor invasion and metastasis (34).
Tumor-associated macrophages perform phenotypic transformation in the TME, driving the TME from an anti-tumor state to an immunosuppressive state, and tumor-associated macrophages are expected to become an important link in the regulation of tumor behavior and efficacy of treatments. In this study, the analysis of immune cell infiltration in primary TNBC tumors and TNBC BMs showed that most immune cells in TNBC BM were significantly increased. The prognosis analysis of TNBC patients without BM showed that the high expression of effector memory CD8 T cells, immature dendritic cells, mast cells, and natural killer cell indicated a poor prognosis, and an increase in immature dendritic cell expression in TNBC BM patients indicated a poor prognosis. Thus, there is a complicated relationship between the presence of BM and the expression of immune cells.
There has been a trend of integration in the treatment of TNBC BM, and drug therapy plays an important role in the treatment of TNBC. Due to the existence of the blood-brain barrier and a lack of sensitive drugs, a number of challenges face the treatment of TNBC BM. In this study, the drug sensitivity analysis showed that TNBC BM patients were more sensitive to Bicalutamide and Imatinib compared with TNBC without BM patients. A series of preclinical and clinical studies have shown that Bicalutamide can improve the prognosis of some androgen antibody-positive patients with metastatic TNBC (35), and Imatinib has a certain effect on radiotherapy and chemotherapy in breast cancer (36). These reports are consistent with the findings of the present study.
Our study had some limitations. First, the TNBC and TNBC BM patients selected from the population were limited in terms of their representativeness, and subsequent studies with large samples need to be conducted. Second, molecular cytology experiments need to be conducted to confirm that the proposed biomarkers of TNBC BM.
Conclusions
AIM2 and ZBP1 are an important pair of pyroptosis genes that can be used as biomarkers for TNBC BM. The expression of AIM2 and ZBP1 may be effective at predicting patients’ responses to treatment and prognosis.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-2182
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-2182). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Bergin ART, Loi S. Triple-negative breast cancer: recent treatment advances. F1000Res 2019; [Crossref] [PubMed]
- Wei W, Cao S, Liu J, et al. Fibroblast growth factor receptor 4 as a prognostic indicator in triple-negative breast cancer. Transl Cancer Res 2020;9:6881-8. [Crossref]
- Qiu J, Xue X, Hu C, et al. Comparison of Clinicopathological Features and Prognosis in Triple-Negative and Non-Triple Negative Breast Cancer. J Cancer 2016;7:167-73. [Crossref] [PubMed]
- Pedrosa RMSM, Mustafa DA, Soffietti R, et al. Breast cancer brain metastasis: molecular mechanisms and directions for treatment. Neuro Oncol 2018;20:1439-49. [Crossref] [PubMed]
- Hosonaga M, Saya H, Arima Y. Molecular and cellular mechanisms underlying brain metastasis of breast cancer. Cancer Metastasis Rev 2020;39:711-20. [Crossref] [PubMed]
- Chang G, Shi L, Ye Y, et al. YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020;38:857-871.e7. [Crossref] [PubMed]
- Cookson BT, Brennan MA. Pro-inflammatory programmed cell death. Trends Microbiol 2001;9:113-4. [Crossref] [PubMed]
- Xia X, Wang X, Cheng Z, et al. The role of pyroptosis in cancer: pro-cancer or pro-"host"? Cell Death Dis 2019;10:650. [Crossref] [PubMed]
- Storr SJ, Safuan S, Ahmad N, et al. Macrophage-derived interleukin-1beta promotes human breast cancer cell migration and lymphatic adhesion in vitro. Cancer Immunol Immunother 2017;66:1287-94. [Crossref] [PubMed]
- Kim MS, Lebron C, Nagpal JK, et al. Methylation of the DFNA5 increases risk of lymph node metastasis in human breast cancer. Biochem Biophys Res Commun 2008;370:38-43. [Crossref] [PubMed]
- Jiao Y, Zhao H, Chen G, et al. Pyroptosis of MCF7 Cells Induced by the Secreted Factors of hUCMSCs. Stem Cells Int 2018;2018:5912194. [Crossref] [PubMed]
- Elion DL, Jacobson ME, Hicks DJ, et al. Therapeutically Active RIG-I Agonist Induces Immunogenic Tumor Cell Killing in Breast Cancers. Cancer Res 2018;78:6183-95. [Crossref] [PubMed]
- Noh MG, Kim SS, Kim YJ, et al. Evolution of the Tumor Microenvironment toward Immune-Suppressive Seclusion during Brain Metastasis of Breast Cancer: Implications for Targeted Therapy. Cancers (Basel) 2021;13:4895. [Crossref] [PubMed]
- Hornung V, Ablasser A, Charrel-Dennis M, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009;458:514-8. [Crossref] [PubMed]
- Man SM, Zhu Q, Zhu L, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell 2015;162:45-58. [Crossref] [PubMed]
- Wilson JE, Petrucelli AS, Chen L, et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med 2015;21:906-13. [Crossref] [PubMed]
- Ma X, Guo P, Qiu Y, et al. Loss of AIM2 expression promotes hepatocarcinoma progression through activation of mTOR-S6K1 pathway. Oncotarget 2016;7:36185-97. [Crossref] [PubMed]
- Martínez-Cardona C, Lozano-Ruiz B, Bachiller V, et al. AIM2 deficiency reduces the development of hepatocellular carcinoma in mice. Int J Cancer 2018;143:2997-3007. [Crossref] [PubMed]
- Li C, Zhang Y, Cheng X, et al. PINK1 and PARK2 Suppress Pancreatic Tumorigenesis through Control of Mitochondrial Iron-Mediated Immunometabolism. Dev Cell 2018;46:441-455.e8. [Crossref] [PubMed]
- Chen IF, Ou-Yang F, Hung JY, et al. AIM2 suppresses human breast cancer cell proliferation in vitro and mammary tumor growth in a mouse model. Mol Cancer Ther 2006;5:1-7. [Crossref] [PubMed]
- Zhang T, Yin C, Boyd DF, et al. Influenza Virus Z-RNAs Induce ZBP1-Mediated Necroptosis. Cell 2020;180:1115-1129.e13. [Crossref] [PubMed]
- Ioannidis P, Trangas T, Dimitriadis E, et al. C-MYC and IGF-II mRNA-binding protein (CRD-BP/IMP-1) in benign and malignant mesenchymal tumors. Int J Cancer 2001;94:480-4. [Crossref] [PubMed]
- Ross J, Lemm I, Berberet B. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene 2001;20:6544-50. [Crossref] [PubMed]
- Ioannidis P, Mahaira L, Papadopoulou A, et al. 8q24 Copy number gains and expression of the c-myc mRNA stabilizing protein CRD-BP in primary breast carcinomas. Int J Cancer 2003;104:54-9. [Crossref] [PubMed]
- Wang W, Goswami S, Lapidus K, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res 2004;64:8585-94. [Crossref] [PubMed]
- Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell 2005;97:97-110. [Crossref] [PubMed]
- Lapidus K, Wyckoff J, Mouneimne G, et al. ZBP1 enhances cell polarity and reduces chemotaxis. J Cell Sci 2007;120:3173-8. [Crossref] [PubMed]
- Gu W, Pan F, Singer RH. Blocking beta-catenin binding to the ZBP1 promoter represses ZBP1 expression, leading to increased proliferation and migration of metastatic breast-cancer cells. J Cell Sci 2009;122:1895-905. [Crossref] [PubMed]
- Gu W, Katz Z, Wu B, et al. Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. J Cell Sci 2012;125:81-91. [Crossref] [PubMed]
- Siveen KS, Kuttan G. Role of macrophages in tumour progression. Immunol Lett 2009;123:97-102. [Crossref] [PubMed]
- Vinogradov S, Warren G, Wei X. Macrophages associated with tumors as potential targets and therapeutic intermediates. Nanomedicine (Lond) 2014;9:695-707. [Crossref] [PubMed]
- Zhang WJ, Wang XH, Gao ST, et al. Tumor-associated macrophages correlate with phenomenon of epithelial-mesenchymal transition and contribute to poor prognosis in triple-negative breast cancer patients. J Surg Res 2018;222:93-101. [Crossref] [PubMed]
- Aras S, Zaidi MR. TAMeless traitors: macrophages in cancer progression and metastasis. Br J Cancer 2017;117:1583-91. [Crossref] [PubMed]
- Gucalp A, Tolaney S, Isakoff SJ, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res 2013;19:5505-12. [Crossref] [PubMed]
- Irie M, Takeuchi Y, Ohtake Y, et al. Imatinib mesylate treatment in a dog with gastrointestinal stromal tumors with a c-kit mutation. J Vet Med Sci 2015;77:1535-9. [Crossref] [PubMed]


