
Identification of key genes involved in tamoxifen-resistant breast cancer using bioinformatics analysis
Introduction
The incidence of breast cancer is increasing in China and in most other countries (1). Breast cancer is the most common malignant neoplasm and the primary cause of cancer-related death in women in the United States (2,3). According to the literature, approximately 70% to 80% of breast cancers express estrogen receptor alpha (ERα) (4,5). Endocrine therapies have significantly reduced the mortality and recurrence rates of ER-positive breast cancer patients. Tamoxifen, a selective estrogen receptor modulator (SERM), has proven effective in the treatment of ER-positive breast cancer in premenopausal women (6). A meta-analysis confirmed that tamoxifen was able to reduce the recurrence rate of breast cancer by approximately 37% and the mortality rate by 29% in women <45 years of age (7); however, about 50% of ER-positive breast cancers present with an inherent resistance to endocrine therapy, and 30% to 40 % of those reactive to tamoxifen will become resistant to tamoxifen (3). The mechanisms of tamoxifen resistance are related to many factors, such as the activation of oncogenes, the inactivation of antioncogenes, changes in the expression of ERα, alterations in co-regulatory proteins, and the influence of growth factor signal pathways (8). Nevertheless, the phenomenon of tamoxifen resistance is still a major clinical problem in breast cancer therapy.
With the application of bioinformatics technology to microarray analysis, significant progress has been made in the study of tamoxifen resistance mechanisms. Some genes have been found to be associated with mechanisms of tamoxifen resistance; for example, the amplified in breast cancer 1 (AIB1) gene, also known as steroid receptor coactivator (SRC-3), is overexpressed in more than 50% of breast cancers (8). AIB1 overexpression correlates with the recurrence of breast tumors, shorter diseasefree survival times, and poorer overall survival (9). Evidence has been provided to show that AIB1 contributes to tamoxifen resistance (10,11). Moreover, a knockdown of AIB1 level in the tamoxifen-resistant breast cancer cell line BT47 was shown to restore the sensitivity of breast cancer cells to tamoxifen (9). Anterior gradient 2 (AGR2), a secretory protein, is a member of the protein disulfide isomerase (PDI) family. It has been discovered that tamoxifen may stimulate the expression of AGR2, and AGR2 overexpression plays a role in tamoxifen resistance (12). Human epidermal growth factor receptor 2 (HER2) and G protein-coupled estrogen receptor 1 (GPER) have been demonstrated to contribute to tamoxifen resistance (10,13). Additionally, the mechanisms of tamoxifen resistance are also related to significant pathways, such as the human epidermal growth factor receptor 2 (HER2) tyrosine kinase pathway and the phosphatidylinositol 3-kinase (PI3K) cell survival pathway (8). Progress has been achieved in illustrating the mechanisms of tamoxifen resistance. Nevertheless, the current knowledge on tamoxifen resistance remains inadequate.
In the present study, a bioinformatics method was applied to analyze gene expression profiles in tamoxifen-resistant breast cancer and to identify the differences between differentially expressed genes (DEGs) in tamoxifen-resistant and tamoxifen-sensitive breast cancer cells. A protein-protein interaction (PPI) network of DEGs was structured for discovering the potential and crucial genes involved in tamoxifen-resistant breast cancer. The purpose of the study was to strengthen the understanding of the mechanisms of tamoxifen resistance and to identify potential novel therapeutic targets for tamoxifen-resistant breast cancer. We present the following article in accordance with the STREGA reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1276).
Methods
Gene expression data analysis
Based on the GPL570 platform data (Affymetrix Human Genome U133 Plus 2.0 Array; Affymetrix, Inc., Santa Clara, CA, USA ), as reported by Elias et al. (14), the GSE67916 gene expression profiles were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo). These data included 4 tamoxifen-resistant MCF7/S0.5 cell lines, TamR1, TamR4, TamR7, and TamR8, and 1 tamoxifen-sensitive MCF7/S0.5 cell line. In total, 18 samples consisting of 9 tamoxifen-resistant breast cancer cell samples and 9 tamoxifen-sensitive breast cancer cell samples were analyzed in the present study.
Statistical analysis
The data were processed and analyzed using R v. 3.6.3 statistical software (R Foundation for Statistical Computing, Vienna, Austria). The average expression value of the probes mapped to the same gene was regarded as the final expression value for the gene.
Data processing and DEG analysis
The DEGs in the tamoxifen-resistant breast cancer cell line and the tamoxifen-sensitive cell line were analyzed using the Linear Models for Microarray Analysis (limma) v. 3.48.3 software package (https://www.bioconductor.org/). Absolute value of log fold change >1.5 and adjusted P<0.01 were considered to be the threshold values for DEGs.
Gene ontology (GO) and pathway enrichment analysis
GO_produced by Gene Ontology Consortium, aims to conveniently and accurately represent the requests for updates to biological information received from scientists making gene and protein annotations (15). In addition, it classifies relevant gene sets into their respective pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/) database.
To analyze the differentially expressed genes, the GO functions and KEGG pathway enrichment analyses were investigated using the Database for Annotation, Visualization and Integration Discovery (DAVID) v. 6.8 software (https://david.abcc.ncifcrf.gov/). GO categories, including molecular function (MF), biological process (BP), and cellular component (CC), were applied to the analysis of the classification of the DEGs. A P value of <0.05 was set as the cutoff criterion.
Construction of the PPI network
The PPI network was established for the DEGs employing the Search Tool for the Retrieval of Interacting Genes (STRING) v. 11 (https://string-db.org), an online database that gathers integrated information of PPIs (16). The interactions of protein pairs in the STRING database were displayed using a combined score, and a combined score of >0.4 was established as the cutoff value in the network. The PPI network was subsequently presented using the Cytoscape v. 3.5.0 software platform, and the hub genes were screened out according to their degree of connectivity in the PPI network (expressed as node degree, referring to number of neighbors). The subnetworks with a node degree >10 were appraised using the Molecular Complex Detection (MCODE) plugin in Cytoscape (17). Subsequently, the subnetwork functions were analyzed by GO and KEGG pathway enrichment analyses using DAVID.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Result
Identification of DEGs
A total of 438 DEGs were identified in the 9 tamoxifen-resistant breast cancer cell samples and the 9 tamoxifen-sensitive breast cancer cell samples using the limma package, of which 300 genes were upregulated and 138 genes were downregulated (Table 1).
Table 1
| Genes | |
|---|---|
| Upregulated | PRKCA, HHLA3, PGM2L1, C8orf44-SGK3///SGK3, C6orf89, MR1, FBP1, SCYL2, CSGALNACT1, FAS, ZDHHC5, MTHFD2, SDC2, EFNA5, CEP97, ENC1, ELOVL2-AS1, EIF2AK2, LMBRD2, RERG, MAP1B, CYB5B, ARHGEF6, SH3BGRL, NUDT7, YPEL2, CLIC3, CLIC4, RHOBTB3, NEK2, USP47, TPBG, SMARCA4, P2RX4, CHRNA5, MYO1B, ZFP36L2, CAB39, POLI, C5orf24, TIA1, KCNJ8, USP42, TMEM87B, GOPC, ZGRF1, KCNK5, ACVR1C, KCTD1, FBXO38, DDX3X, GPATCH2, C12orf65, ASAH1, SQSTM1, NME7, ENTPD3, CTDSPL2, ACTA2, MIR612///NEAT1, BTN3A2, CTSO, CTNNA1, TRIM38, DGKH, SPTAN1, OSMR, NKTR, BBX, CDK13, GLUD2, TEX9, FARSA, RAB32, TMC5, KIAA0513, ONECUT2, BHLHE41, SLC1A4, SELM, MBNL1, HNRNPD, WDR90, SPAG9, LOC101928524, SETBP1, LMNB1, TMPRSS3, AGFG1, PARP9, LOC642852, TPR, GALC, VAV3, HIST1H2AM, NR2F2, CPEB4, FOXO3B///FOXO3, CYP4B1, HIST1H2AE, GADD45B, RAB5A, TRIM6-TRIM34///TRIM34, ARHGAP5, ARID1A, ID2B///ID2, SLC7A8, JUND, RBAK, MINCR, NR1D2, HLA-DQB1, AKAP1, SGMS2, IGF1R, FAM76A, PIK3R1, RECQL4, FUS, DLX2, MIA3, LOC101060835///LOC100996809///HLA-DRB5///HLA-DRB4///HLA-DRB1///HLA-DQB1, CAPN8, FAM117A, FDFT1, F11R, ZNF431, MT1F, NUP210, ASAP1, TM7SF2, SP110, SDC1, SYTL2, TFPI, ST8SIA4, SALL4, SNX27, GALNT2, CDKL5, KIF5C, DCAF16, SNORA28///EIF5, GNB4, FREM2, LGALS3BP, ABCD3, ZNF850, EXPH5, EHD1, TMX1, ATP6V1A, LIPA, GBP2, MAP2, DGKA, OGDH, ADGRL2, TRIO, STIP1, NFIB, MAP3K1, PCDH19, PMEPA1, FAM213A, MAN1A1, AMOTL1, HIST2H4B///HIST4H4///HIST2H4A///HIST1H4L///HIST1H4E///HIST1H4B///HIST1H4H///HIST1H4C///HIST1H4J///HIST1H4K///HIST1H4F///HIST1H4D///HIST1H4A///HIST1H4I, C1orf115, TIAM1, FNIP1, ZYX, RBM22, GATC, CLCC1, HSPA2, U2SURP, SP100, LOC100288911, MVD, GDPD1, MAVS, SMTN, PHF10, C17orf80, EDEM3, LYST, PPP2R3B, SLC24A3, PAPOLA, ADCY1, RANBP2, DACH1, TFPI2, BCAT1, HDAC8, ZNF253, MYCBP2, TRIM59///IFT80, GGCX, PGGT1B, HMGN5, ANXA9, SLC22A15, LOC101929597///TBCD, HIPK1, KPNA1, NBPF1, CFL1, GSAP, WTAP, TPP1, GBP3, MYEF2, DENND4C, FNIP2, SCUBE2, KRIT1, CCDC83, GULP1, 6-Mar, SUGT1, FYTTD1, CLDN3, NDRG1, STX17, EFNB2, C21orf91, KDM4B, C14orf132, ATXN1, CENPF, MSMO1, LY6E, ACYP1, AKAP13, ERVMER34-1, AGA, ANLN, EIF5A2, GOSR2, KLHL20, ANOS1, RBM48, R3HCC1L, STC2, CITED1, GGT6, SECISBP2L, NDST4, EXOC6, RUNDC1, USP10, JPX, USP34, BTN3A2///BTN3A3, GEM, SRSF4, SLC4A7, BBS9, ITGB4, XPO7, AGR2, TANK, IRF7, CITED2, CACNG4, CYP1B1, PRNP, NR3C1, KRT15, SERPINI1, MAPK1IP1L, PHF23, CEBPG, GRN, ZMYM2, METTL8, CA5BP1, DNMT3A, TANC2, MPPED2, HERPUD2, PABPN1, TBL1X, TMEFF2, TBRG1, RAB30, ZNF711, SEC61A1, LAMP1, SYNRG, BRINP3, ZYG11B, MBTPS1, SRRT, ZNRF2, SEPHS1, FBXW11, PHLDA1 |
| Downregulated | SHANK2, CPSF6, MOCS3, C1GALT1, TOB2, WBSCR22, AP1S3, WLS, METTL4, DEGS1, MED30, MAPKAP1, HLA-A, RBM26, NCOA3, SLC39A6, SLC9A7, CD163L1, EPHA4, GFPT1, SNORD3D///SNORD3C///SNORD3B-2///SNORD3A///SNORD3B-1, BFSP1, KIF3A, TFRC, PRKAB1, ARHGEF28, UBE2J1, PLA2G16, ZNF652, SLC18B1, IFNGR1, CCDC88C, RAD52, FARSB, CAPRIN1, FRMD5, CSNK1A1, MIR1908///FADS1, MRPL30, SETD5, SCPEP1, TIMP2, FAM83A, ATP1B1, LZTFL1, ZNF236, LOC101060835///HLA-DQB1, COL12A1, CA8, LINC00839, CHTF18, GPX3, PSME4, MBP, GPR87, HIST1H4H, PM20D2, SERPINA1, PSEN1, KIFAP3, SYTL4, FGFR2, ERAP1, SGK1, LSS, SOX2, FSIP1, TM9SF1, PCYOX1, TFF1, IRF9, ARFIP1, ZFP90, RAP2A, RBMS1, LARP4, LOC101930578///TPTE2P2, RAMP1, ESR1, MPZL2, KBTBD2, ZNFX1, ELL2, SEPT2, SLC44A1, S100A4, EPC1, MYB, COBLL1, HHIPL2, B4GALT1, HSPA4, FLVCR2, SLC12A6, CA12, LOC100505984///ITGB6, SCN1B, JPH1, LNPEP, SOWAHC, KCTD6, PPM1E, PAPSS2, ATP10D, CDC14B, TBC1D3P1-DHX40P1///RNFT1, PJA1, ZNF703, JMY, MAPK1, FAM189A2, SHMT2, CD59, ARSG, HIST1H2BD, MIR4784///MZT2A///MZT2B, MGP, DLC1, STK38, CPNE4, SOX3, C1orf226, SPANXA2///SPANXA1, TMEM192, HOOK3, CXCL12, PGR, PIK3R3, MALRD1, SMAD5, WWP1, IL20, GBP1, ELOVL2, GFRA1, PLXDC2, GREB1, FCMR |
438 DEGs indentified between tamoxifen resistant and tamoxifen sensitive samples are listed, of which 300 upregulated and 138 downregulated. DEGs, differentially expressed genes.
GO function enrichment analyses of upregulated and downregulated DEGs
GO functional analyses were carried out on the upregulated and downregulated DEGs, respectively. The top 5 GO terms identified in the 3 GO categories (BP, CC, and MF) are shown in Table 1. The GO terms significantly involved in the GO categories of upregulated DEGs were cytoplasm, cytosol, and perinuclear region of cytoplasm in the CC category; protein binding, poly(A) RNA binding, and transcription factor binding in the MF category; and protein transport, negative regulation of transcription from RNA polymerase II promoter, and transcription, DNA-templated in the BP category. The GO terms significantly involved in the GO categories of downregulated DEGs were plasma membrane and extracellular exosome in the CC category; protein binding, kinase activity, and cadherin binding involved in cell-cell adhesion in the MF category; and cellular response to estradiol stimulus, positive regulation of transcription, DNA template, and interferon gamma–mediated signaling pathway in the BP category.
KEGG pathway enrichment analyses of upregulated and downregulated DEGs
The upregulated DEGs were significantly enriched in the leukocyte transendothelial migration, lysosome, and RIG-I-like receptor signaling pathways. The downregulated DEGs were significantly enriched in the viral carcinogenesis pathways, the signaling pathways regulating the pluripotency of stem cells, and the HIF-1 signaling pathway (Table 2).
Table 2
| Category | Term | Count | P value |
|---|---|---|---|
| Upregulated DEGs | |||
| GOTERM_CC_DIRECT | GO:0005737-cytoplasm | 98 | 4.79×10−3 |
| GOTERM_CC_DIRECT | GO:0005829-cytosol | 69 | 2.19×10−3 |
| GOTERM_CC_DIRECT | GO:0016020-membrane | 54 | 2.05×10−4 |
| GOTERM_CC_DIRECT | GO:0048471-perinuclear region of cytoplasm | 19 | 5.18×10−3 |
| GOTERM_CC_DIRECT | GO:0043231-intracellular membrane-bounded organelle | 17 | 8.95×10−3 |
| GOTERM_MF_DIRECT | GO:0005515-protein binding | 159 | 1.69×10−3 |
| GOTERM_MF_DIRECT | GO:0005524-ATP binding | 34 | 2.04×10−2 |
| GOTERM_MF_DIRECT | GO:0044822-poly(A) RNA binding | 26 | 3.98×10−2 |
| GOTERM_MF_DIRECT | GO:0000166-nucleotide binding | 13 | 7.41×10−3 |
| GOTERM_MF_DIRECT | GO:0008134-transcription factor binding | 11 | 1.22×10−2 |
| GOTERM_BP_DIRECT | GO:0000122-negative regulation of transcription from RNA polymerase II promoter | 20 | 1.45×10−2 |
| GOTERM_BP_DIRECT | GO:0045892-negative regulation of transcription, DNA-templated | 15 | 2.12×10−2 |
| GOTERM_BP_DIRECT | GO:0015031-protein transport | 12 | 4.04×10−2 |
| GOTERM_BP_DIRECT | GO:0007399-nervous system development | 10 | 3.23×10−2 |
| GOTERM_BP_DIRECT | GO:0051260-protein homooligomerization | 8 | 1.89×10−2 |
| KEGG_PATHWAY | hsa05164:Influenza A | 11 | 6.44×10−4 |
| KEGG_PATHWAY | hsa05161:Hepatitis B | 7 | 3.48×10−2 |
| KEGG_PATHWAY | hsa04670:Leukocyte transendothelial migration | 7 | 1.41×10−2 |
| KEGG_PATHWAY | hsa04142:Lysosome | 7 | 1.58×10−2 |
| KEGG_PATHWAY | hsa04622:RIG-I-like receptor signaling pathway | 5 | 2.96×10−2 |
| Downregulated DEGs | |||
| GOTERM_CC_DIRECT | GO:0005886-plasma membrane | 37 | 4.59×10−2 |
| GOTERM_CC_DIRECT | GO:0070062-extracellular exosome | 36 | 1.36×10−4 |
| GOTERM_CC_DIRECT | GO:0005829-cytosol | 31 | 4.64×10−2 |
| GOTERM_CC_DIRECT | GO:0005654-nucleoplasm | 28 | 2.77×10−2 |
| GOTERM_CC_DIRECT | GO:0016020-membrane | 25 | 1.04×10−2 |
| GOTERM_MF_DIRECT | GO:0005515-protein binding | 80 | 1.03×10−3 |
| GOTERM_MF_DIRECT | GO:0005524-ATP binding | 15 | 1.67×10−2 |
| GOTERM_MF_DIRECT | GO:0042802-identical protein binding | 12 | 1.67×10−2 |
| GOTERM_MF_DIRECT | GO:0016301-kinase activity | 5 | 9.09×10−3 |
| GOTERM_MF_DIRECT | GO:0098641-cadherin binding involved in cell-cell adhesion | 4 | 3.35×10−2 |
| GOTERM_BP_DIRECT | GO:0045893-positive regulation of transcription, DNA-templated | 10 | 1.10×10−2 |
| GOTERM_BP_DIRECT | GO:0060333-interferon-gamma-mediated signaling pathway | 4 | 1.39×10−2 |
| GOTERM_BP_DIRECT | GO:0006730-one-carbon metabolic process | 3 | 1.90×10−2 |
| GOTERM_BP_DIRECT | GO:0071392-cellular response to estradiol stimulus | 3 | 2.03×10−2 |
| GOTERM_BP_DIRECT | GO:0060348-bone development | 3 | 3.57×10−2 |
| KEGG_PATHWAY | hsa04919:Thyroid hormone signaling pathway | 6 | 2.56×10−3 |
| KEGG_PATHWAY | hsa05203:Viral carcinogenesis | 6 | 2.81×10−2 |
| KEGG_PATHWAY | hsa04550:Signaling pathways regulating pluripotency of stem cells | 5 | 2.95×10−2 |
| KEGG_PATHWAY | hsa04960:Aldosterone-regulated sodium reabsorption | 4 | 4.14×10−3 |
| KEGG_PATHWAY | hsa04066:HIF-1 signaling pathway | 4 | 4.87×10−2 |
Top five GO terms in diversetables and major pathways are listed. Count, number of DEGs; GO,gene ontology; DEGs, differentially expressed genes; MF, molecular function; BP, biological process; CC, cellular component; KEGG, KyotoEncyclopedia of Genes and Genomes.
PPI network analysis
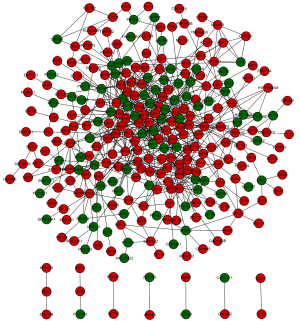
Based on the STRING database analysis, a total of 629 protein pairs with combined scores of >0.4 were identified. As demonstrated in Figure 1, the PPI network consisted of 288 nodes and 629 edges. The nodes of MAPK1 (node degree 36), ESR1 (node degree 27), SMARCA4 (node degree 27), RANBP2 (node degree 25), and PRKCA (node degree 21) were hub proteins in the PPI network.
Two subnetworks (subnetworks 1 and 2) with >10 nodes were discerned using the MCODE plugin (Figure 2). The hub proteins MAPK1 and ESR1 were demonstrated to be involved in subnetwork 1. Subnetwork 1 was primarily associated with the following GO terms: protein binding, DNA binding, nucleoplasm, immune response, positive regulation of transcription, and DNA template. Pathway analysis showed enrichment of the viral carcinogenesis and cancer pathways (Table 3). In contrast, subnetwork 2 was associated with the following GO terms: nucleus, extracellular exosome, and the binding with protein and DNA (Table 4). The most significant pathway in subnetwork 2 was the MAPK signaling pathway (Table 4).
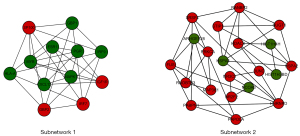
Table 3
| Category | Term | Count | P value |
|---|---|---|---|
| GOTERM_CC_DIRECT | GO:0005654-nucleoplasm | 6 | 1.71×10−2 |
| GOTERM_CC_DIRECT | GO:0000139-Golgi membrane | 3 | 4.76×10−2 |
| GOTERM_MF_DIRECT | GO:0005515-protein binding | 11 | 8.43×10−3 |
| GOTERM_MF_DIRECT | GO:0003677-DNA binding | 6 | 2.64×10−3 |
| GOTERM_MF_DIRECT | GO:0042802-identical protein binding | 5 | 9.88×10−4 |
| GOTERM_MF_DIRECT | GO:0019899-enzyme binding | 3 | 1.89×10−2 |
| GOTERM_MF_DIRECT | GO:0001077-transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | 3 | 9.85×10−3 |
| GOTERM_BP_DIRECT | GO:0060333-interferon-gamma-mediated signaling pathway | 6 | 5.30×10−10 |
| GOTERM_BP_DIRECT | GO:0060337-type I interferon signaling pathway | 5 | 6.21×10−8 |
| GOTERM_BP_DIRECT | GO:0045893-positive regulation of transcription, DNA-templated | 5 | 2.43×10−4 |
| GOTERM_BP_DIRECT | GO:0006955-immune response | 4 | 2.22×10−3 |
| GOTERM_BP_DIRECT | GO:0006366-transcription from RNA polymerase II promoter | 4 | 3.89×10−3 |
| KEGG_PATHWAY | hsa05203:Viral carcinogenesis | 5 | 8.43×10−5 |
| KEGG_PATHWAY | hsa05168:Herpes simplex infection | 4 | 1.36×10−3 |
| KEGG_PATHWAY | hsa04914:Progesterone-mediated oocyte maturation | 3 | 5.33×10−3 |
| KEGG_PATHWAY | hsa04114:Oocyte meiosis | 3 | 8.26×10−3 |
| KEGG_PATHWAY | hsa05200:Pathways in cancer | 3 | 8.91×10−3 |
Top five GO terms in diversetables and major pathways are listed. Count, number of DEGs; GO,gene ontology; DEGs, differentially expressed genes; MF, molecular function; BP, biological process; CC, cellular component; KEGG, KyotoEncyclopedia of Genes and Genomes.
Table 4
| Category | Term | Count | P value |
|---|---|---|---|
| GOTERM_CC_DIRECT | GO:0005634-nucleus | 17 | 3.84×10−5 |
| GOTERM_CC_DIRECT | GO:0005654-nucleoplasm | 11 | 7.49×10−2 |
| GOTERM_CC_DIRECT | GO:0070062-extracellular exosome | 10 | 3.58×10−3 |
| GOTERM_CC_DIRECT | GO:0000790-nuclear chromatin | 4 | 1.55×10−3 |
| GOTERM_CC_DIRECT | GO:0000786-nucleosome | 4 | 1.91×10−4 |
| GOTERM_MF_DIRECT | GO:0005515-protein binding | 20 | 8.13×10−4 |
| GOTERM_MF_DIRECT | GO:0003723-RNA binding | 7 | 5.39×10−5 |
| GOTERM_MF_DIRECT | GO:0003677-DNA binding | 7 | 1.75×10−2 |
| GOTERM_MF_DIRECT | GO:0008270-zinc ion binding | 6 | 1.54×10−2 |
| GOTERM_MF_DIRECT | GO:0044822-poly(A) RNA binding | 6 | 1.34×10−2 |
| GOTERM_MF_DIRECT | GO:0019899-enzyme binding | 4 | 8.87×10−3 |
| GOTERM_BP_DIRECT | GO:0000398-mRNA splicing, via spliceosome | 5 | 1.80×10−4 |
| GOTERM_BP_DIRECT | GO:0031124-mRNA 3’-end processing | 3 | 1.93×10−3 |
| GOTERM_BP_DIRECT | GO:0006369-termination of RNA polymerase II transcription | 3 | 3.14×10−3 |
| KEGG_PATHWAY | hsa05034:Alcoholism | 5 | 9.61×10−4 |
| KEGG_PATHWAY | hsa05322:Systemic lupus erythematosus | 4 | 4.70×10−3 |
| KEGG_PATHWAY | hsa04010:MAPK signaling pathway | 4 | 2.69×10−2 |
Top five GO terms in diversetables and major pathways are listed. Count, number of DEGs; GO, geneontology; MF, molecular function; BP, biological process; CC, cellular component; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
Tamoxifen acts as a first-line therapy in the treatment of ER-positive breast cancer for premenopausal women. Yet, the phenomenon of tamoxifen resistance has become a major clinical problem in breast cancer therapy. It is therefore critical that novel therapeutic targets for tamoxifen-resistant breast cancer are discovered. To date, many relevant proteins and pathways have been identified. The receptor tyrosine kinase (RTK) family and activation of the phosphatidylinositol-3-kinase (PI3K)/AKT/mammalian target of the rapamycin (mTOR) receptor pathway are thought to be important mechanisms of tamoxifen resistance (18). In recent years, it has been proven that low expression of ERα36 increases the tamoxifen sensitivity of breast cancer cells by blocking the epidermal growth factor receptor/extracellular signal-regulated kinase (EGFR/ERK) signaling pathway (19). Gao et al. found that the LEM4 structural protein inhibits the sensitivity of breast cancer cells to tamoxifen by accelerating the G1 to S phase (G1/S) transition (20). In addition, it is reported that highly expressed cell division cycle–associated protein 8 (CDCA8) may increase tamoxifen resistance in breast cancer cells (21). Elias et al. (14) reported that several functional genes, including FYN, PRKCA, ITPR1, DPYD, DACH1, LYN, GBP1, and PRLR, are related to a reduction in tamoxifen sensitivity. In the present study, we reanalyzed the updated data. A total of 438 DEGs in tamoxifen-resistant and tamoxifen-sensitive breast cancer cell-lines were identified, including 300 upregulated and 138 downregulated genes. GO function and KEGG pathway enrichment analysis were used to analyze the DEGs, which were associated with protein binding and immune response. In addition, signaling pathway analysis revealed that these DEGs were mainly involved in the MAPK signaling pathway in cancer. Subsequently, the PPI network and subnetworks were constructed in order to explore the interactions of the DEGs. The top t nodes (by maximum node degree, referring to number of neighbors) were identified, including MAPK1, ESR1, SMARCA4, RANBP2, and PRKCA.
MAPK1, also known as ERK, encodes the protein which is involved in a wide variety of cellular processes, such as proliferation, differentiation, transcription regulation, and development (22,23).With the phosphorylation of 90 kDa ribosomal S6 kinases (p90RSK2), ERK mediates mitogen-induced proliferation signals from the cell membrane to the nucleus (24). MAPK1 mutations are associated with many human cancers, such as breast cancer, prostate cancer, and ovarian cancer (23,25,26). Furthermore, some studies have proven that MAPK1 hyperactivation plays a role in tamoxifen resistance (27,28). ERα is phosphorylated by increased ERK activity, which leads to a ligand-independent transcription of ERα and an agonistic activity of tamoxifen (29). The present study indicated that MAPK1 was a hub protein with a node degree score of 36 in the PPI network. Therefore, the MAPK1 gene may be a crucial regulator in tamoxifen-resistant breast cancer.
SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 (SMARCA4), also known as brahma-related gene-1 (BRG1), codes a core protein of the SWI/SNF chromatin-remodeling complex, which impacts chromatin and the transcription of target genes via the energy of ATP hydrolysis. Moreover, it also controls many cellular processes, such as DNA repair (30). BRG1 mutations are found in lung cancer and Burkitt’s lymphoma (31,32). High BRG1 expression has been associated with poor survival and cell proliferation of triple-negative breast cancer (TNBC) (33). Furthermore, prior reports have indicated that BRG1 is related to estrogen receptor and is recruited to estrogen-responsive promoters (34). Therefore, BRG1 may play a role in the mechanism of estrogen antagonists. Nacht et al. demonstrated the function of BRG1 in hormone-dependent gene repression in breast cancer cells. BRG1 plays a key role in hormone-dependent cell proliferation and apoptosis (35). In addition, SMARCA4 was observed acting as a potential regulator of differentially expressed proteins in male breast cancer (36); however, the linkage between BRG1 and tamoxifen resistance is rarely reported. A later mechanistic study revealed that SOX4 and SMARCA4 cooperatively regulate PI3K/Akt signaling and lead to the genesis and/or progression of TNBC (37). In our study, the SMARCA4 gene was elevated in the tamoxifen-resistant samples and was a hub protein in the PPI network. Thus, SMARCA4 may be a potential target in the treatment of tamoxifen-resistant breast cancer.
The ESR1 gene has been found to encode the ERα and a ligand-dependent transcription factor of the nuclear receptor family (38). ERα can regulate the activities of genes in various biological and tumor progression processes, and plays a key role in endocrine therapies for ER-positive breast cancer (39). Significant research efforts have demonstrated that the loss of ERα expression or function may contribute to resistance to tamoxifen therapy (8). The ligand-independent activity of ERα mutants may mediate resistance to tamoxifen (38,40). In accordance with the findings of the present study, decreased ESR1 expression was reported in tamoxifen-resistant samples of a previous study by Kim et al. (41), and ESR1 was found to act as a hub gene in the PPI network. All of these findings suggest that ESR1 has a central role in resistance to tamoxifen.
The RAN-binding protein 2 (RANBP2) is located at chromosomal region 2q13 and was initially considered to be a regulated factor of nucleo-cytoplasmic trafficking (42). RANBP2 encodes a nucleoporin with 358-kDa that functions in nuclear export or import, including mitotic progression (43,44). Felix et al. reported that, compared to plasma cells, the overexpression of RANBP2 was found in >50% of multiple myeloma cases (45). In addition, the gene is implicated in some tumorigenic pathways, for example, indirectly in the p53 and PI3K/Akt pathways (46,47). Recently, there has been increasing evidence to show that the upregulation of RANBP2 promotes cancer cell growth in cholangiocarcinoma, cervical cancer, and hepatocellular carcinoma (48-50). From the present study, it is evident that RANBP2 serving as an upregulated gene may impact tamoxifen resistance. Due to the function of RANBP2 in cancer, it is likely to become a viable therapeutic approach for treating tamoxifen resistance.
The protein kinase C alpha (PRKCA) gene, one of the protein kinase C (PKC) family members, is activated by a variety of stimuli, including tyrosine kinase receptors and guanine nucleotide-binding protein-coupled receptors, and plays critical roles in many different cellular processes, including the cellular functions of proliferation, apoptosis, and differentiation (51). Kim et al. (52) indicated that PKC-α mediated cell invasion and migration in breast cancer cells. Moreover, it was found that PKC-α could inhibit ER-α expression by suppressing c-Jun phosphorylation and that the level of PKC-α phosphorylation was significantly increased in the tamoxifen-resistant cell line (41). These data imply that PKC-α is a potential biomarker in tamoxifen resistance.
Conclusions
A total of 438 DEGs were revealed in tamoxifen-resistant breast cancer and tamoxifen-sensitive samples using gene expression profiles. Among these DEGs, MAPK1, ESR1, SMARCA4, RANBP2, and PRKCA were found to act as hub genes, and they may participate in the important biological processes and pathways involved in the mechanism of tamoxifen resistance. Further research, however, is required to validate these potential therapies.
Acknowledgments
First and foremost, we extend our appreciation to Professor Jun Liu for her guidance and discussions. Second, we would like to thank Professor Qing Chen, as we would not have been able to continue this study without her patience and encouragement.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1276
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-1276
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1276). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang H, Tan G, Dong L, et al. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One 2012;7:e34210. [Crossref] [PubMed]
- Li G, Zhang J, Jin K, et al. Estrogen receptor-α36 is involved in development of acquired tamoxifen resistance via regulating the growth status switch in breast cancer cells. Mol Oncol 2013;7:611-24. [Crossref] [PubMed]
- Vanderlaag KE, Hudak S, Bald L, et al. Anterior gradient-2 plays a critical role in breast cancer cell growth and survival by modulating cyclin D1, estrogen receptor-alpha and survivin. Breast Cancer Res 2010;12:R32. [Crossref] [PubMed]
- Kazi AA, Gilani RA, Schech AJ, et al. Nonhypoxic regulation and role of hypoxia-inducible factor 1 in aromatase inhibitor resistant breast cancer. Breast Cancer Res 2014;16:R15. [Crossref] [PubMed]
- Al-Dhaheri M, Wu J, Skliris GP, et al. CARM1 is an important determinant of ERα-dependent breast cancer cell differentiation and proliferation in breast cancer cells. Cancer Res 2011;71:2118-28. [Crossref] [PubMed]
- Ignatov A, Ignatov T, Roessner A, et al. Role of GPR30 in the mechanisms of tamoxifen resistance in breast cancer MCF-7 cells. Breast Cancer Res Treat 2010;123:87-96. [Crossref] [PubMed]
- Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 2011;378:771-84. [Crossref] [PubMed]
- Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer 2004;11:643-58. [Crossref] [PubMed]
- Chang AK, Wu H. The role of AIB1 in breast cancer. Oncol Lett 2012;4:588-94. [Crossref] [PubMed]
- Su Q, Hu S, Gao H, et al. Role of AIB1 for tamoxifen resistance in estrogen receptor-positive breast cancer cells. Oncology 2008;75:159-68. [Crossref] [PubMed]
- Karmakar S, Foster EA, Blackmore JK, et al. Distinctive functions of p160 steroid receptor coactivators in proliferation of an estrogen-independent, tamoxifen-resistant breast cancer cell line. Endocr Relat Cancer 2011;18:113-27. [Crossref] [PubMed]
- Brychtova V, Vojtesek B, Hrstka R. Anterior gradient 2: a novel player in tumor cell biology. Cancer Lett 2011;304:1-7. [Crossref] [PubMed]
- Ignatov A, Ignatov T, Weissenborn C, et al. G-protein-coupled estrogen receptor GPR30 and tamoxifen resistance in breast cancer. Breast Cancer Res Treat 2011;128:457-66. [Crossref] [PubMed]
- Elias D, Vever H, Lænkholm AV, et al. Gene expression profiling identifies FYN as an important molecule in tamoxifen resistance and a predictor of early recurrence in patients treated with endocrine therapy. Oncogene 2015;34:1919-27. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Liu X, Ma Y, Yang W, et al. Identification of therapeutic targets for breast cancer using biological informatics methods. Mol Med Rep 2015;12:1789-95. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Yao J, Deng K, Huang J, et al. Progress in the Understanding of the Mechanism of Tamoxifen Resistance in Breast Cancer. Front Pharmacol 2020;11:592912. [Crossref] [PubMed]
- Li G, Zhang J, Xu Z, et al. ERα36 as a Potential Therapeutic Target for Tamoxifen-Resistant Breast Cancer Cell Line Through EGFR/ERK Signaling Pathway. Cancer Manag Res 2020;12:265-75. [Crossref] [PubMed]
- Gao A, Sun T, Ma G, et al. LEM4 confers tamoxifen resistance to breast cancer cells by activating cyclin D-CDK4/6-Rb and ERα pathway. Nat Commun 2018;9:4180. [Crossref] [PubMed]
- Yu D, Shi L, Bu Y, et al. Cell Division Cycle Associated 8 Is a Key Regulator of Tamoxifen Resistance in Breast Cancer. J Breast Cancer 2019;22:237-47. [Crossref] [PubMed]
- Elloumi-Mseddi J, Jemel-Oualha I, Beji A, et al. Effect of estradiol and clomiphene citrate on Erk activation in breast cancer cells. J Recept Signal Transduct Res 2015;35:202-6. [Crossref] [PubMed]
- Jung YC, Han S, Hua L, et al. Kazinol-E is a specific inhibitor of ERK that suppresses the enrichment of a breast cancer stem-like cell population. Biochem Biophys Res Commun 2016;470:294-9. [Crossref] [PubMed]
- Lee CJ, Lee HS, Ryu HW, et al. Targeting of magnolin on ERKs inhibits Ras/ERKs/RSK2-signaling-mediated neoplastic cell transformation. Carcinogenesis 2014;35:432-41. [Crossref] [PubMed]
- Chen QG, Zhou W, Han T, et al. MiR-378 suppresses prostate cancer cell growth through downregulation of MAPK1 in vitro and in vivo. Tumour Biol 2016;37:2095-103. [Crossref] [PubMed]
- Zou Y, Deng W, Wang F, et al. A novel somatic MAPK1 mutation in primary ovarian mixed germ cell tumors. Oncol Rep 2016;35:725-30. [Crossref] [PubMed]
- Kronblad A, Hedenfalk I, Nilsson E, et al. ERK1/2 inhibition increases antiestrogen treatment efficacy by interfering with hypoxia-induced downregulation of ERalpha: a combination therapy potentially targeting hypoxic and dormant tumor cells. Oncogene 2005;24:6835-41. [Crossref] [PubMed]
- Generali D, Buffa FM, Berruti A, et al. Phosphorylated ERalpha, HIF-1alpha, and MAPK signaling as predictors of primary endocrine treatment response and resistance in patients with breast cancer. J Clin Oncol 2009;27:227-34. [Crossref] [PubMed]
- de Leeuw R, Neefjes J, Michalides R. A role for estrogen receptor phosphorylation in the resistance to tamoxifen. Int J Breast Cancer 2011;2011:232435. [Crossref] [PubMed]
- Wu JI. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim Biophys Sin (Shanghai) 2012;44:54-69. [Crossref] [PubMed]
- Rodriguez-Nieto S, Cañada A, Pros E, et al. Massive parallel DNA pyrosequencing analysis of the tumor suppressor BRG1/SMARCA4 in lung primary tumors. Hum Mutat 2011;32:E1999-2017. [Crossref] [PubMed]
- Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 2012;44:1321-5. [Crossref] [PubMed]
- Wu Q, Madany P, Akech J, et al. The SWI/SNF ATPases Are Required for Triple Negative Breast Cancer Cell Proliferation. J Cell Physiol 2015;230:2683-94. [Crossref] [PubMed]
- Wang S, Zhang B, Faller DV. BRG1/BRM and prohibitin are required for growth suppression by estrogen antagonists. EMBO J 2004;23:2293-303. [Crossref] [PubMed]
- Nacht AS, Beato M, Vicent GP. Steroid hormone receptors silence genes by a chromatin-targeted mechanism similar to those used for gene activation. Transcription 2017;8:15-20. [Crossref] [PubMed]
- Gomig THB, Gontarski AM, Cavalli IJ, et al. Integrated analysis of label-free quantitative proteomics and bioinformatics reveal insights into signaling pathways in male breast cancer. Genet Mol Biol 2021;44:e20190410. [Crossref] [PubMed]
- Mehta GA, Angus SP, Khella CA, et al. SOX4 and SMARCA4 cooperatively regulate PI3k signaling through transcriptional activation of TGFBR2. NPJ Breast Cancer 2021;7:40. [Crossref] [PubMed]
- Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al. D538G mutation in estrogen receptor-α: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res 2013;73:6856-64. [Crossref] [PubMed]
- Jeselsohn R, Buchwalter G, De Angelis C, et al. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol 2015;12:573-83. [Crossref] [PubMed]
- Jeselsohn R, Yelensky R, Buchwalter G, et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res 2014;20:1757-67. [Crossref] [PubMed]
- Kim S, Lee J, Lee SK, et al. Protein kinase C-α downregulates estrogen receptor-α by suppressing c-Jun phosphorylation in estrogen receptor-positive breast cancer cells. Oncol Rep 2014;31:1423-8. [Crossref] [PubMed]
- Hashizume C, Kobayashi A, Wong RW. Down-modulation of nucleoporin RanBP2/Nup358 impaired chromosomal alignment and induced mitotic catastrophe. Cell Death Dis 2013;4:e854. [Crossref] [PubMed]
- Klein UR, Haindl M, Nigg EA, et al. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell 2009;20:410-8. [Crossref] [PubMed]
- Lee SE, Kang SY, Takeuchi K, et al. Identification of RANBP2-ALK fusion in ALK positive diffuse large B-cell lymphoma. Hematol Oncol 2014;32:221-4. [Crossref] [PubMed]
- Felix RS, Colleoni GW, Caballero OL, et al. SAGE analysis highlights the importance of p53csv, ddx5, mapkapk2 and ranbp2 to multiple myeloma tumorigenesis. Cancer Lett 2009;278:41-8. [Crossref] [PubMed]
- Miyauchi Y, Yogosawa S, Honda R, et al. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J Biol Chem 2002;277:50131-6. [Crossref] [PubMed]
- Packham S, Warsito D, Lin Y, et al. Nuclear translocation of IGF-1R via p150(Glued) and an importin-β/RanBP2-dependent pathway in cancer cells. Oncogene 2015;34:2227-38. [Crossref] [PubMed]
- Yang J, Liu Y, Wang B, et al. Sumoylation in p27kip1 via RanBP2 promotes cancer cell growth in cholangiocarcinoma cell line QBC939. BMC Mol Biol 2017;18:23. [Crossref] [PubMed]
- Wang H, Luo Q, Kang J, et al. YTHDF1 Aggravates the Progression of Cervical Cancer Through m6A-Mediated Up-Regulation of RANBP2. Front Oncol 2021;11:650383. [Crossref] [PubMed]
- Liu X, Chen X, Xiao M, et al. RANBP2 Activates O-GlcNAcylation through Inducing CEBPα-Dependent OGA Downregulation to Promote Hepatocellular Carcinoma Malignant Phenotypes. Cancers (Basel) 2021;13:3475. [Crossref] [PubMed]
- Nakashima S. Protein kinase C alpha (PKC alpha): regulation and biological function. J Biochem 2002;132:669-75. [Crossref] [PubMed]
- Kim S, Han J, Lee SK, et al. Berberine suppresses the TPA-induced MMP-1 and MMP-9 expressions through the inhibition of PKC-α in breast cancer cells. J Surg Res 2012;176:e21-9. [Crossref] [PubMed]
(English Language Editors: K. Gilbert and J. Gray)


