
Association between single nucleotide polymorphisms of IL-6 and susceptibility to skin cancer: a meta-analysis and systematic review
Introduction
Skin cancer is one of the most common malignant cancers in the world, which is often caused by ultraviolet radiation, immunosuppressive therapy or radiotherapy (1). Skin cancer can be classified according to the origin of malignant cells, which are usually located in the epidermis, dermis or skin appendages. Malignant cancers derived from the epidermis include cutaneous melanoma (CM) and non-melanoma skin cancers (NMSCs), such as basal cell carcinoma (BCC), squamous cell carcinoma (SCC) and Merkel cell carcinoma (MCC) (2). BCC and SCC are the most common types of skin cancer, which account for about 80% and 16% of all NMSCs, respectively (3). Melanoma accounts for only 10% of skin cancer, but melanoma is the leading cause of death due to skin cancer (4). At present, although there is considerable treatment progress worldwide, skin cancers are heterogenic and present with diverse malignant potential, prognosis and treatment methods.
Several cytokines, such as interleukin-6 (IL-6), are known to inhibit the proliferation of melanoma cell lines and regulate the invasive features of melanoma cells (5). Abnormal IL-6 levels might be associated with the occurrence of skin cancer (6), and has been found to exert an important inhibition effect on melanoma cell growth (7). IL-6 overexpression was confirmed in primary cutaneous lymphoma (8). The levels of STAT3 and IL-6 are also higher in patients with autoimmune diseases (9), in which, IL-6 not only maintains inflammation, but also regulates immune response. It can be considered that IL-6 can regulate the shift of immune response by changing the quality of peptide presentation, so as to activate or tolerate (10). Thus, the gene polymorphisms of cytokines might be associated with the susceptibility to skin cancer.
A number of studies have investigated the relationship of IL-6 single nucleotide polymorphisms (SNPs), including rs1800795-174G/C and rs1800797-597G/A, with disease susceptibility and prognosis. A few literatures found out that polymorphisms of IL-6 might affect the occurrence of skin cancer (11), while several studies suggested the opposite (12). For instance, Sławińska et al. found that the C allele in rs1800795-174 was correlated with significantly increased risk of BCC (C vs. G: OR =1.86, P=0.004) (13). On the contrary, Wang et al. discovered that the G allele in rs1800795-174 might enhance the susceptibility to BCC (G vs. C: OR =1.38, P=0.012) (14). Five years earlier, a meta-analysis by Wu et al. summarized and analyzed the association between IL-6 polymorphism and skin cancer, and found that rs1800795-174G/C was associated with the risk of skin cancer (for GC vs. GG: OR =1.28, P=0.816; for CC/GC vs. GG: OR =1.26, P=0.618) (15). However, the data in the meta-analysis by Wang et al. could not draw a definite conclusion, and the included literatures were 10 years ago. In the recent decade, several new reports that investigate the association between IL-6 gene SNP and skin cancer risk have been published. A new meta-analysis is needed to further verify this question. In addition, another SNP (597G/A) of IL-6 has not been fully studied due to the lack of previous literatures. Therefore, this meta-analysis also aims to explore the relationship between two IL-6 SNPs and the risk of skin cancer.
We present the following article in accordance with the PRISMA reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1508).
Methods
Database search
We searched all papers focusing on the relationship between IL-6 174G/C and 597G/A polymorphisms and skin cancer through July 2021 in the PubMed, Embase, Web of Science and Cochrane. The keywords were used as following:
- For IL-6 polymorphism: IL-6, interleukin-6, rs1800795, 174G/C, rs1800797 and 597G/A;
- For polymorphism: polymorphism, variant and SNP;
- For skin cancer: skin cancer, basal cell carcinoma, cutaneous squamous cell carcinoma, BCC, SCC and melanoma.
The electronic search was supplemented by examining the reference lists of other articles identified and reviewing original report. Endnotes (version X7) was used to manage search records for literature screening. The protocol of this meta-analysis has been registered in the International Prospective Register of Systematic Reviews (PROSPERO, registration ID: CRD42021271467). The eligibility of identified literatures was independently determined by two authors, and the divergences in the results were resolved through discussion with the other author. All human-related studies were included if the following criteria were met: (I) case-control study; (II) the outcome was skin cancer; (III) relevant genotype data were available to calculate the odds ratios (ORs) with 95% confidence intervals (CIs). The major exclusion criteria were: (I) duplicate data; (II) review, meta-analysis or guideline; (III) case report, letter, comment, editorial, protocol, reply, etc.; (IV) with less than 20 patients; (V) basic research; (VI) non-relevant topic; (VII) article not in English; (VIII) no available data were reported (Figure 1).
Data extraction
According to the inclusion and exclusion criteria listed above, eligible data was extracted by two authors independently from every study. The authors discussed to settle the different result in data extraction. If the authors fails to reach an agreement, the third author will attend the negotiation to resolve the dispute and make the final decision through a majority vote. The following data were extracted: name of the first author, year of publication, genotyping method, region of study, cancer type, the numbers of patients and controls, and the distribution of genotypes in case and control groups.
Assessment of study quality
For the included studies, the Newcastle-Ottawa scale (NOS) was calculated for quality assessment. If the research score is 8 or 9, the research quality is considered to be high; If the study score is 6 or 7, the study is considered to be of medium quality; If the study score is <6, the study is considered to be of poor quality.
Statistical analysis
Deviation of frequencies of IL-6 gene -174G/C and 597G/A polymorphisms from Hardy-Weinberg equilibrium (HWE) was tested by Chi-square test in cases and controls, and P<0.05 was considered as significant. ORs and 95% CI were used to measure the related strength of IL-6 174G/C and 597G/A polymorphisms and the skin cancer. The Z test determined the significance of OR, and P<0.05 was considered statistically significant. Subgroup analysis was carried out according to publication year, cancer type, sample size, and NOS score. I2 statistic test was performed to examine the heterogeneity of the studies, and P<0.05 was considered significant. Due to the substantial heterogeneity found in the included studies, we used the random effects model to analyze the cumulative incidence rate via the package of meta in R version 4.0.2 (https://www.r-project.org/).
Publication bias was estimated by using funnel-plot and Egger’s test. Trim-and-fill method aiming to adjust OR value was performed when publication bias was found. By using the meta package in R, sensitivity analysis was carried out through omitting each study successively.
Results
Characteristics of included studies
The flow chart of study selection is depicted in Figure 1. A total of 378 articles regarding IL-6 gene 174G/C and 597G/A polymorphisms and skin cancer were recognized. One hundred and twenty-eight articles were excluded because due to duplicates, and 187 articles were excluded due to the type of publication, sample size and non-relevant topic. In addition, 5 publications were excluded due to the language other than English. Another 47 publications were excluded due to no available data. Finally, 11 studies were included in the meta-analysis (5,11-14,16-21). For each included study, we evaluated the NOS score to estimate the literature quality.
As shown in Table 1, 11 publications included 1,705 cases and 1,987 controls for IL-6 174G/C polymorphism (10 publications), and 968 cases and 998 controls IL-6 gene 597G/A polymorphism (3 publications). For IL-6 174G/C polymorphism, there were 5 melanoma studies, 4 BCC studies, and 1 cutaneous T-cell lymphoma (CTCL) study. Meanwhile there were 1 melanoma studies and 2 BCC studies for IL-6 597G/A polymorphism. The detailed genotype distribution is listed in Table 2. The distributions of genotypes in the case group and control group were in accordance with the Hardy-Weinberg balance except for 2 studies (Table 2).
Table 1
| Author | Year | Region | Cancer type | No. of patients | No. of controls | Genotype method | NOS score |
|---|---|---|---|---|---|---|---|
| Martínez-Escribano | 2002 | Spain | Melanoma | 42 | 48 | PCR-SSP | 7 |
| Howell | 2003 | UK | Melanoma | 161 | 224 | ARMS-PCR | 8 |
| Zhang | 2004 | Sweden | BCC | 241 | 260 | Pyrosequencing | 8 |
| Vasku | 2004 | Czech Republic | CTCL | 63 | 105 | PCR-RFLP | 6 |
| Nikolova | 2007 | Bulgaria | Melanoma | 120 | 120 | PCR-SSP | 6 |
| Wilkening | 2006 | Germany | BCC | 529 | 533 | TaqMan | 7 |
| Vogel | 2007 | Denmark | BCC | 304 | 315 | TaqMan | 9 |
| Gu | 2008 | USA | Melanoma | 207 | 204 | TaqMan | 7 |
| Sławińska | 2019 | Poland | BCC | 243 | 198 | PCR-SSP | 8 |
| Vlaykova | 2020 | Bulgaria | Melanoma | 59 | 173 | PCR-RFLP | 8 |
| Wang | 2020 | China | BCC | 265 | 340 | PCR-SSP | 9 |
NOS, Newcastle-Ottawa scale; BCC, basal cell carcinoma; CTCL, cutaneous T-cell lymphoma; PCR-SSP, polymerase chain reaction with sequence-specific primers; ARMS-PCR, amplification refractory mutation system-polymerase chain reaction; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
Table 2
| Author | 174G/C-case | PHWE | 174G/C-control | PHWE | 597G/A-case | PHWE | 597G/A-control | PHWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GC | CC | GG | GC | CC | GG | GA | AA | GG | GA | AA | |||||
| Martínez-Escribano | 14 | 26 | 2 | 0.078 | 20 | 26 | 2 | 0.2 | NA | NA | NA | NA | NA | NA | NA | NA |
| Howell | 48 | 79 | 34 | 0.99 | 79 | 101 | 44 | 0.53 | NA | NA | NA | NA | NA | NA | NA | NA |
| Zhang | 57 | 126 | 58 | 0.778 | 62 | 130 | 68 | 1 | 59 | 112 | 70 | 0.567 | 68 | 123 | 69 | 0.686 |
| Vasku | 19 | 35 | 9 | 0.54 | 36 | 46 | 23 | 0.53 | NA | NA | NA | NA | NA | NA | NA | NA |
| Nikolova | 35 | 56 | 29 | 0.78 | 51 | 55 | 14 | 0.99 | NA | NA | NA | NA | NA | NA | NA | NA |
| Wilkening | NA | NA | NA | NA | NA | NA | NA | NA | 207 | 226 | 96 | 0.051 | 171 | 270 | 92 | 0.709 |
| Vogel | 65 | 176 | 63 | 0.023 | 89 | 157 | 69 | 1 | NA | NA | NA | NA | NA | NA | NA | NA |
| Gu | 69 | 106 | 32 | 0.706 | 69 | 102 | 33 | 0.9 | 28 | 102 | 68 | 0.58 | 34 | 100 | 71 | 0.993 |
| Sławińska | 96 | 91 | 56 | 0.002 | 93 | 74 | 31 | 0.052 | NA | NA | NA | NA | NA | NA | NA | NA |
| Vlaykova | 30 | 22 | 7 | 0.651 | 74 | 83 | 16 | 0.57 | NA | NA | NA | NA | NA | NA | NA | NA |
| Wang | 28 | 117 | 120 | 0.998 | 23 | 131 | 186 | 1 | NA | NA | NA | NA | NA | NA | NA | NA |
HWE, Hardy-Weinberg equilibrium; NA, not available.
Quantitative synthesis
The assessments of the relationship of 174G/C and 597G/A polymorphisms and skin cancer risk are shown in Figures 2,3, respectively. When all eligible studies were included in analysis, no elevated risk of skin cancer was found in all genotypes for 174G/C (Figure 2):


CC vs. GC + GG, OR =1.03 (95% CI: 0.81–1.31);
GC + CC vs. GG, OR =1.16 (95% CI: 0.96–1.39);
CC vs. GG, OR =1.14 (95% CI: 0.86–1.53);
GC vs. GG, OR =1.16 (95% CI: 0.99–1.37);
C vs. G, OR =1.07 (95% CI: 0.92–1.24).
Similarly, no significant risk of skin cancer was found in all genotypes for 597G/A (Figure 3):
GG vs. GA + AA, OR =1.07 (95% CI: 0.77–1.49);
GA + GG vs. AA, OR =0.94 (95% CI: 0.76–1.16);
GG vs. AA, OR =1.01 (95% CI: 0.78–1.30);
GA vs. AA, OR =0.89 (95% CI: 0.71–1.12);
G vs. A, OR =1.03 (95% CI: 0.90–1.18).
Due to the limited number of studies for 597G/A (n=3), the following subgroup analysis, sensitivity analysis and publication bias were only evaluated for 174G/C.
Subgroup analysis
We performed subgroup analysis based on publication year, the cancer type, the subtypes of non-melanoma (BCC and CTCL), sample size and NOS score. As shown in Table 3, the analysis results of the subtypes of non-melanoma group were non-significant. BCC is the most common NMSC, and our result indicated there was no association between IL-6 gene -174GC polymorphism and BCC. However, due to the limited research included in our study, this conclusion still needs to be confirmed by future research. In conclusion, none of the subgroups showed meaningful results except the subgroup of publication year before 2010 (GC + CC vs. GG: OR =1.255, P=0.012; GC vs. GG: OR =1.277, P=0.01), which is consistent with the previous meta-analysis by Wu et al. (15). On the contrary, there is no statistical significance in the other subgroup of publication year after 2010 (P>0.05 for all genotypes). Thus, publication date might also be one of the sources of heterogeneity.
Table 3
| Subgroup | No. of studies | CC vs. GC + GG | GC + CC vs. GG | CC vs. GG | GC vs. GG | C vs. G | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | P value | OR | P value | OR | P value | OR | P value | OR | P value | ||||||
| Total | 10 | 1.03 | 0.818 | 1.16 | 0.126 | 1.14 | 0.363 | 1.16 | 0.066 | 1.07 | 0.4 | ||||
| Publication year | |||||||||||||||
| Before 2010 | 7 | 1.02 | 0.875 | 1.255 | 0.012 | 1.198 | 0.221 | 1.277 | 0.01 | 1.109 | 0.135 | ||||
| After 2010 | 3 | 1.086 | 0.801 | 0.878 | 0.629 | 1.003 | 0.994 | 0.882 | 0.531 | 0.96 | 0.855 | ||||
| Type of cancer | |||||||||||||||
| Melanoma | 5 | 1.257 | 0.19 | 1.178 | 0.258 | 1.381 | 0.134 | 1.125 | 0.373 | 1.157 | 0.189 | ||||
| Non-melanoma | 5 | 0.906 | 0.522 | 1.127 | 0.391 | 0.997 | 0.989 | 1.189 | 0.143 | 1.003 | 0.973 | ||||
| BCC | 4 | 0.949 | 0.755 | 1.106 | 0.548 | 1.036 | 0.88 | 1.153 | 0.305 | 1.016 | 0.904 | ||||
| CTCL | 1 | 0.594 | 0.227 | 1.208 | 0.581 | 0.741 | 0.537 | 1.442 | 0.312 | 0.931 | 0.754 | ||||
| Sample size | |||||||||||||||
| <300 | 4 | 1.262 | 0.52 | 1.218 | 0.361 | 1.416 | 0.35 | 1.168 | 0.461 | 1.159 | 0.395 | ||||
| ≥300 | 6 | 0.958 | 0.718 | 1.132 | 0.252 | 1.065 | 0.688 | 1.166 | 0.095 | 1.031 | 0.724 | ||||
| NOS score | |||||||||||||||
| ≤7 | 4 | 1.131 | 0.708 | 1.266 | 0.094 | 1.321 | 0.433 | 1.249 | 0.134 | 1.175 | 0.284 | ||||
| >7 | 6 | 0.982 | 0.893 | 1.082 | 0.539 | 1.082 | 0.635 | 1.1 | 0.435 | 1.021 | 0.826 | ||||
IL-6, interleukin-6; OR, odds ratio; BCC, basal cell carcinoma; CTCL, cutaneous T-cell lymphoma; NOS, Newcastle-Ottawa scale.
Sensitivity analysis
As shown in Figure 4, for all comparisons of 174G/C genotypes, OR estimates (ranging from 1.03 to 1.16) showed well stability within the 95% confidence interval of the combined OR. The OR estimates of CC vs. GC + GG, GC + CC vs. GG, CC vs. GG, GC vs. GG, and C vs. G, remained stable when omitting one study at each analysis. Most estimates showed no statistically significance (P>0.05).

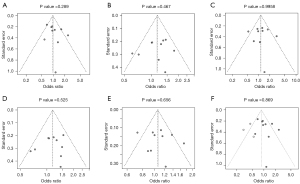
Publication bias
In the five comparisons of 174G/C genotypes, Egger’s test did not reveal significant publication bias (P>0.05 for all). Funnel plots are shown in Figure 5. Slight asymmetry was found in the funnel of CC vs. GC + GG (Figure 5A). Therefore, we carried out the trim-and-fill method to evaluate the missing literatures, so as to make the OR value more reliable. After the trim-and-fill adjustment, three miss literatures were added (Figure 5F), and the OR value was further decreased (OR =0.855; 95% CI: 0.647–1.128; P=0.268). This further supports the conclusion that IL-6 gene 174G/C polymorphism might not be associated with the risk of skin cancer.
Discussion
The incidence rate of melanoma and NMSCs has increased worldwide in recent decades. Epidemiological and experimental evidence shows that inflammation is one of the signs of skin cancer induced by solar ultraviolet radiation and several other environmental factors (3). Inflammatory cytokines play a considerable role in the initial stage of tumorigenesis, so as to influence the risk of cancer. IL-6 is a pleiotropic cytokine produced by multiple cell types in a variety of tissues, which has been proven to regulate the immune defense mechanism by initiating the acute phase of immune response (2,22). In addition to being an effective immunomodulator, it can also affect the growth of transformed cells (10). The polymorphism of cytokine gene regulatory region is related to the amount of cytokine produced in vitro, and the synthesis of IL-6 has been proved to be related to the genotype variation at position 174G/C (23-25). Gene polymorphisms can exert a pathophysiological effect role by regulating the host’s immune response to skin cells. By regulating the level of gene expression as well as its function, polymorphisms of immune related genes could have direct functional significance.
Recently, increasing studies have investigated the relationship between IL-6 gene 174G/C and 597G/A polymorphisms and skin cancer risk, yet the results remain controversial. In order to get a comprehensive conclusion, we performed this meta-analysis which include a total of 11 studies. Overall, we found that the risk of skin cancer was not related with both IL-6 gene polymorphisms. The earlier meta-analysis by Wu et al. found the IL-6 gene 174G/C might be associated with increased skin cancer risk (15), which is contradictory as compared with ours. We considered that the reasons for the different results may be due to the newly published studies. The included literatures in the meta-analysis by Wu et al. were before 2010 (15), and the number of literatures and patients included were less. Meanwhile, the newly included literatures in our meta-analysis were published within recent 3 years, which is far from the age of previous literatures. Therefore, the selection criteria of patients and the control of confounding factors in the literatures can be quite different. For instance, the control groups of our newly included literature were all healthy controls, but several literatures in previous years did not mention whether the source of the control was healthy individuals or patients with other diseases. On the other hand, the NOS score of the newly included literature is evaluated as high quality (8 and 9 points), while a proportion of literatures in previous years is relatively lower (<8 points). These factors may lead to the different results between ours and those previously published by Wu et al.
The relationship between IL-6 polymorphisms and skin cancer risk has been under debate. Zhang et al. supported the conclusion that IL-6 gene 597G/A polymorphism was not related with an increased risk of skin cancer (16), meanwhile Vogel et al. also suggested that IL-6 gene 174G/C polymorphism did not increase the risk of skin cancer (19). In addition, the most recent two studies by Vlaykova and Wang, respectively, both indicated that the allele G might be associated with a higher risk of skin cancer, which is to the past views the patients with allele C are at hazard of melanoma and BCC (14,21). Furthermore, when evaluating the publication bias, we used the trim-and-fill method to estimate the missing unpublished literatures. Instead, the OR value after adjustment was less than 1, indicating that there might be publication bias of the literatures in previous years. Taken together, the existing evidence suggests that there might not be significant correlation between IL-6 gene 174G/C polymorphism and skin cancer.
597G/A is another SNP located in the promoter region of IL-6, which can regulate the immune response by regulating the level of IL-6 in vivo (16). There are few studies on this SNP in the past, and some studies have found 597G/A was related with an increase risk of skin cancer (16,18), while the other suggested the opposite (20). Our meta-analysis included three articles which had data on the relationship between 597G/A and skin cancer. The raw data were pooled and analyzed, but no significant association was found. The number of literatures included was relatively small, which limited the power of data analysis, thus the relationship between IL-6 gene 597G/A and skin cancer needs to be verified by more future studies.
Despite the efforts we made to adjust for potential bias and to elucidate the relationship between IL-6 gene 174G/C and 597G/A polymorphisms and skin cancer, several limitations existed in the study. First, current available data could not further clarify the inter-reaction between SNPs and environment based on the aggregate data. Secondly, although comprehensive literatures were searched in PubMed, Embase, Web of Science and Cochrane, which have no language restrictions, the funnel plots still suggested that there might be publication bias in terms of the association of IL-6 gene 174G/C polymorphism and skin cancer, and the positive results were more likely to be published as compared with negative results and inconclusive results. Despite all this, we performed trim-and-fill method to reduce this type of bias and obtained more credible results. Third, the sample size is not large enough, which limits the statistical power, especially for the studies of 597G/A polymorphisms. More studies, especially large-scale prospective studies, are still required to further explore the relationship between IL-6 gene 174G/C and 597G/A polymorphisms and skin cancer.
Conclusions
Overall, the results in this meta-analysis showed that IL-6 gene polymorphisms might not be associated with the risk of skin cancer. In the future, large-scale and well-designed studies are needed to further address this issue.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1508
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1508). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schierbeck J, Vestergaard T, Bygum A. Skin Cancer Associated Genodermatoses: A Literature Review. Acta Derm Venereol 2019;99:360-9. [Crossref] [PubMed]
- Catalano O, Roldán FA, Varelli C, et al. Skin cancer: findings and role of high-resolution ultrasound. J Ultrasound 2019;22:423-31. [Crossref] [PubMed]
- Maru GB, Gandhi K, Ramchandani A, et al. The role of inflammation in skin cancer. Adv Exp Med Biol 2014;816:437-69. [Crossref] [PubMed]
- Garbe C, Peris K, Hauschild A, et al. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline--Update 2012. Eur J Cancer 2012;48:2375-90. [Crossref] [PubMed]
- Martínez-Escribano JA, Moya-Quiles MR, Muro M, et al. Interleukin-10, interleukin-6 and interferon-gamma gene polymorphisms in melanoma patients. Melanoma Res 2002;12:465-9. [Crossref] [PubMed]
- Weber R, Riester Z, Hüser L, et al. IL-6 regulates CCR5 expression and immunosuppressive capacity of MDSC in murine melanoma. J Immunother Cancer 2020;8:e000949. [Crossref] [PubMed]
- Krasagakis K, Garbe C, Orfanos CE. Cytokines in human melanoma cells: synthesis, autocrine stimulation and regulatory functions--an overview. Melanoma Res 1993;3:425-33. [Crossref] [PubMed]
- Asadullah K, Gellrich S, Haeussler-Quade A, et al. Cytokine expression in primary cutaneous germinal center cell lymphomas. Exp Dermatol 2000;9:71-6. [Crossref] [PubMed]
- Eriksen KW, Kaltoft K, Mikkelsen G, et al. Constitutive STAT3-activation in Sezary syndrome: tyrphostin AG490 inhibits STAT3-activation, interleukin-2 receptor expression and growth of leukemic Sezary cells. Leukemia 2001;15:787-93. [Crossref] [PubMed]
- Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev 2002;13:357-68. [Crossref] [PubMed]
- Vasku JA, Vasku A, Goldbergova M, et al. Heterozygote AG variant of -596 A/G IL-6 gene polymorphism is a marker for cutaneous T-cell lymphoma (CTCL). Clin Immunol 2004;113:256-60. [Crossref] [PubMed]
- Howell WM, Turner SJ, Theaker JM, et al. Cytokine gene single nucleotide polymorphisms and susceptibility to and prognosis in cutaneous malignant melanoma. Eur J Immunogenet 2003;30:409-14. [Crossref] [PubMed]
- Sławińska M, Zabłotna M, Gleń J, et al. STAT3 polymorphisms and IL-6 polymorphism are associated with the risk of basal cell carcinoma in patients from northern Poland. Arch Dermatol Res 2019;311:697-704. [Crossref] [PubMed]
- Wang J, Chen Y. Interleukin-6 -174 G/C polymorphism is associated with the risk of basal cell carcinoma in a Chinese Han population. Aging (Albany NY) 2020;12:15328-33. [Crossref] [PubMed]
- Wu M, Cao Y, Liu YL, et al. Meta-analysis of the Correlation Between Interleukin-6 Promoter Polymorphism -174G/C and Interferon Regulatory Factor 4 rs12203592 Polymorphism With Skin Cancer Susceptibility. Am J Ther 2016;23:e1758-67. [Crossref] [PubMed]
- Zhang Z, Liu W, Jia X, et al. Use of pyrosequencing to detect clinically relevant polymorphisms of genes in basal cell carcinoma. Clin Chim Acta 2004;342:137-43. [Crossref] [PubMed]
- Nikolova PN, Pawelec GP, Mihailova SM, et al. Association of cytokine gene polymorphisms with malignant melanoma in Caucasian population. Cancer Immunol Immunother 2007;56:371-9. [Crossref] [PubMed]
- Wilkening S, Hemminki K, Rudnai P, et al. Case-control study in basal cell carcinoma of the skin: single nucleotide polymorphisms in three interleukin promoters pre-analysed in pooled DNA. Br J Dermatol 2006;155:1139-44. [Crossref] [PubMed]
- Vogel U, Christensen J, Wallin H, et al. Polymorphisms in COX-2, NSAID use and risk of basal cell carcinoma in a prospective study of Danes. Mutat Res 2007;617:138-46. [Crossref] [PubMed]
- Gu F, Qureshi AA, Niu T, et al. Interleukin and interleukin receptor gene polymorphisms and susceptibility to melanoma. Melanoma Res 2008;18:330-5. [Crossref] [PubMed]
- Vlaykova T, Kurzawski M, Tacheva T, et al. Effects of the IL6 -174G>C promoter polymorphism and IL-6 serum levels on the progression of cutaneous malignant melanoma. Oncol Lett 2020;20:1781-91. [Crossref] [PubMed]
- Ramadori G, Christ B. Cytokines and the hepatic acute-phase response. Semin Liver Dis 1999;19:141-55. [Crossref] [PubMed]
- Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest 1998;102:1369-76. [Crossref] [PubMed]
- Perrey C, Pravica V, Sinnott PJ, et al. Genotyping for polymorphisms in interferon-gamma, interleukin-10, transforming growth factor-beta 1 and tumour necrosis factor-alpha genes: a technical report. Transpl Immunol 1998;6:193-7. [Crossref] [PubMed]
- Pravica V, Asderakis A, Perrey C, et al. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-gamma gene. Eur J Immunogenet 1999;26:1-3. [Crossref] [PubMed]



