
Functional clustering analysis identifies specific subtypes of aldehyde dehydrogenase associated with glioma immunity
Introduction
Gliomas are one of the most aggressive and infiltrative tumors in the central nervous system (CNS). The 5-year survival rate of patients with glioblastoma multiforme (GBM), which is classified as a grade IV glioma by the World Health Organization (WHO), is less than 5% (1). Therefore, new effective therapeutic methods for gliomas are an expanding focus of research, including immunotherapy and small molecule inhibitors therapy (2,3). Isocitrate dehydrogenase (IDH) mutation, which involves the metabolism of cancer cells, is one of the most important mutations of gliomas (4). Clinical and basic research about IDH mutation has made encouraging progress, such as identification of the inhibitor of IDH (5). Emerging evidence has shown that 2-hydroxyglutarate (2-HG), a metabolite of mutated IDH, contributes to the inhibitory immune microenvironment in glioblastoma (6). Research has implied that distinct subtypes of metabolic enzymes may have crucial roles in the crosstalk between tumors and their immune microenvironments, rather than acting independently within the tumor.
Aldehyde dehydrogenases (ALDH enzymes), as oxidizing enzymes which are composed of 19 isoforms, play a crucial part in cell proliferation, differentiation, metabolism, and other functions, and recent evidence has suggested that a high expression of ALDH is related to cancer malignancy (7). Moreover, mounting evidence shows that ALDH, particularly ALDH1A3, has close ties to tumorigenesis and tumor progression (8-10). Interrupting the ALDH metabolic pathway has become a promising therapeutic strategy. However, side effects induced by the inhibition of ALDH enzymes remain unclear. We hypothesized that a target specific ALDH isozyme would cause an increased expression of one or several ALDH isozymes for functional compensation. At present, classification of ALDH enzymes is mainly based on the molecular structure of the enzymes, but this has not been confirmed in clinical application (11). Therefore, reclassification of ALDH isoforms based on their functions is urgently needed.
In this study, we developed a new approach for the reclassification of ALDH enzymes. We divided ALDH enzymes into 5 subgroups based on their functional evaluation across 2 databases, the Chinese Glioma Genome Atlas (CGGA) and The Cancer Genome Atlas (TCGA). These 5 subgroups had different characteristics, and all ALDH enzymes in the same subgroup had similar functions. Importantly, we identified 3 subgroups of ALDH enzymes that relate to tumor immune functions. This new classification displayed the biological functions of different ALDH enzymes and could improve the clinical application of ALDH inhibitors in glioma treatment. This reclassification process enabled us to better understand the functions and characteristics of ALDH enzymes in gliomas. Furthermore, our study provided a theoretical basis for ALDH functional research and ALDH targeted immunotherapy of glioma patients. We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) checklist (available at https://dx.doi.org/10.21037/tcr-21-1160).
Methods
This study included messenger RNA (mRNA) sequencing data from 999 patients with gliomas of all grades. The sequencing data of the test group were from the CGGA. The sequencing was performed with the Illumina Hiseq 2000 platform (Illumina, San Diego, CA, USA). Only samples that consisted of more than 80% of tumor cells were used. Overall survival (OS) was calculated from the date of diagnosis to death or the last follow-up. Transcriptome sequencing data and patient information can be freely downloaded from the CGGA portal (http://www.cgga.org.cn). The sequencing data of the validation group were from TCGA mRNA-seq database downloaded from public databases (https://cancergenome.nih.gov). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Tiantan Hospital, Capital Medical University (KY2020-093-02). The institutional review board decided that informed consent was not required.
Functional clustering analysis
The biological function and signal pathway activation scores of each participant were calculated by gene set variation analysis (GSVA). The calculation process selected default settings. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets were downloaded from gene set enrichment analysis (GSEA) web portals (http://software.broadinstitute.org/gsea/index.jsp). Subsequently, the correlation between biological function scores and gene expressions was calculated using Pearson correlation analysis. A matrix of correlation coefficients of all patients with 4,653 GO terms and 186 KEGG terms was obtained. Finally, functional clustering results were acquired using unsupervised cluster analysis based on the data above. The integrated clustering analysis was performed by Cluster-of-Cluster Assignments analysis as previously described (12). All statistical computations and figure drawing were performed with the statistical software environment R version 3.5.0 (http://www.r-project.org/).
Chromosome stability and DNA repair functions
The chromosome stability was evaluated by aneuploidy scores, which reflect the alterations of chromosome arm-level copy-numbers in a sample (13). This score can be further divided into chromosome arm-level loss and gain scores. The GSVA scores of DNA repair gene set evaluated DNA repair functions. The relationship between ALDH enzymes and the above functions was analyzed using Pearson correlation analysis. Furthermore, the correlation analyses between ALDH enzymes and leukocyte, tumor purity, mutation number, and stemness index were also performed with the same method.
Immune functions analysis
The relationship between ALDH enzymes and immune functions was evaluated using the Pearson correlation analysis of ALDH enzymes’ mRNA expression and immune function scores. Immune function scores were calculated using GSVA analysis, and the immune function gene set was downloaded from AmiGO 2 web portals (http://amigo.geneontology.org/amigo/landing). The classification of immune functions was based on the classification in the AmiGO2 website.
Prognostic analysis
The prognostic value of ALDH enzymes or ALDH groups was estimated by Kaplan-Meier analysis or Cox analysis. Kaplan-Meier analysis was performed with the statistical software environment R version 3.5.0, and Cox analysis was performed with the statistical software SPSS 25.0 (IBM Corp., Armonk, NY, USA). The median was used as the cutoff of each group in all prognostic analyses. When more than 2 genes were involved in the grouping, each gene group’s intersection was used as a grouping standard.
Statistical analysis
All statistical computations and figure drawing were performed with R packages (ggplot, pheatmap, and survival), GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA), and Microsoft Office 2016 pro (Microsoft, Redmond, WA, USA). Statistical differences between 2 populations were calculated byt-test. The log-rank test was used to assess the statistical significance of stratified survival groups. For all statistical methods, P<0.05 was considered a significant difference.
Results
A new classification of ALDH enzymes based on their biological functions and signaling activation
To reclassify ALDH isoforms based on genome, we performed unsupervised cluster analysis on their mRNA expression level and gene methylation level in the CGGA database (Figure 1A,1B). However, no significant classification trend was observed. A similar situation was seen in TCGA database (Figure S1A,S1B). Therefore, we developed a new approach to reclassify ALDH enzymes based on their functions. When using the new functional clustering method (details in methods), the isoforms of ALDH enzymes could be divided into 5 groups according to their biological functions (GO groups 1–5) and signal pathway activations (KEGG groups 1–5), respectively (Figure 1C,1D; https://cdn.amegroups.cn/static/public/tcr-21-1160-1.xlsx). Subsequently, we saw similar results in the integrated cluster analysis (Figure 1E). As expected, functional clustering analysis also worked well in TCGA database (Figure S1C-S1E). Notably, the classification results were highly consistent in both databases.

Different ALDH enzymes in the same subgroup most likely play similar biological roles in glioma
The landscape of the biological functions of ALDH enzymes revealed that each group has its specific functional spectrum in CGGA (Figure 2; https://cdn.amegroups.cn/static/public/tcr-21-1160-2.xlsx) and TCGA databases (Figure S2; https://cdn.amegroups.cn/static/public/tcr-21-1160-3.xlsx). In Group I, all 3 isoforms were positively correlated with most GO terms (>50%) related to behavior and transportation. Meanwhile, they were significantly negatively correlated with most immune system processes, proliferation, and cell cycle, response to stimulus, and transcription and translation GO terms. Like Group I, ALDH enzymes in Group II were also negatively related to the majority of death and apoptosis, proliferation and cell cycle, response to stimulus and transcription, and translation GO terms, but did not show a positive relationship with any biological function category. Group III and ALDH enzymes were highly positively correlated with modification, transcription, and translation, and negatively correlated with the immune system process. Unlike other groups, ALDH enzymes in Group IV were positively correlated with most immune system processes, signaling, and response to stimulus GO terms. Nevertheless, for Group V, the situation was different; ALDH enzymes showed almost no specific functions.

ALDH functional subgroups are related to clinical characteristics in glioma patients
To explore the relationship between clinical features and our new classification, we analyzed the ALDH expression of each isoform in different pathological grades based on the 2021 WHO Classification of Tumors of the Central Nervous System. Compared to WHO II grade gliomas, the expression of ALDH enzymes in Group I and Group II was significantly lower than the expression in WHO III, but all ALDH enzymes in Group IV showed the opposite trend. Moreover, a similar situation was observed in Group I and Group IV between WHO III and WHO IV, while ALDH enzymes in other Groups showed no apparent difference (Figure 3A-3E; Figure S3). Univariate Cox analysis was performed to further explore the prognostic values of ALDH enzymes in glioma. Consistent with the above clinical results, high expression of ALDH isoforms in Group I and Group II was a more effective prognostic indicator, and high expression in Group IV was a less favorable prognostic indicator (Figure 3F). In summary, we found that ALDH enzymes, especially those in Group I, Group II, and Group IV, were closely related to the clinical phenotype in glioma patients.

ALDH functional subgroups are related to chromosome stability and DNA repair function
Gene mutation and stability of chromosomes are vital factors that affect patient prognosis. Further studies were performed to verify whether ALDH enzymes in each group were associated with gene mutations (https://cdn.amegroups.cn/static/public/tcr-21-1160-4.xlsx). The results showed that total mutation counts were significantly lower in the higher expression isoforms of Group I and Group II and significantly higher in the higher expression isoforms of Group IV. No such difference was observed in Groups III and V (Figure 4A). As for the chromosome stability, the trends of differences were consistent with total mutations, whether for chromosome amplification or deletion (Figure 4B,4C). To further explore the causes of DNA mutation, we measured the relationship between all ALDH enzymes and the DNA repair function scores through the correlation coefficient. We found that 2 kinds of DNA repair functions were most positively related to the expression of ALDH enzymes in Group I and Group II and negatively related to the expression of ALDH enzymes in Group IV.

Conversely, a DNA repair function was negatively related to ALDH isoforms in Group I or Group II and positively related to the expression of ALDH isoforms in Group IV (Figure 4D; Figure S4A). Altogether, we demonstrated that ALDH enzymes, particularly those in Group I, II, and IV, were closely related to gene mutation. The critical factor in this process may be DNA repair functions.
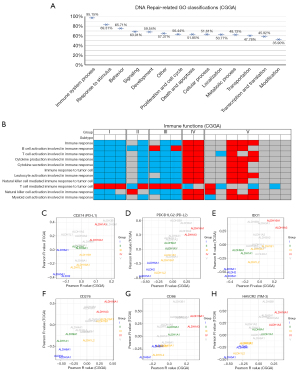
ALDH functional subgroups are closely related to the tumor immune microenvironment
The function of DNA repair is likely to be a crucial reason for the malignant process of glioma, but how it influences the biological function is still unknown. To investigate this, we assessed the correlation between DNA-repair functions and GO terms in CGGA and TCGA databases (Figure 5A; Figure S4B). We found that more than 95% of DNA repair functions had a strong connection with the immune system processes, much more than any other kind of GO term. To understand the differences in immune status between different groups, we performed a correlation coefficient analysis in CGGA and TCGA databases (Figure 5A; Figure S4C). We observed that most immune functions, except for the T cell-mediated immune response to tumor cell (T cell response), were negatively correlated with most ALDH isoforms in Group I, II, and III (Figure 5B). However, the situation in Group IV was different. Most of the immune functions except for T cell response had a positive correlation with ALDH1A3 and ALDH16A1. This abnormal phenomenon made us think of the immune depletion in tumors caused by immune checkpoints. We investigated the correlation between all ALDH enzymes and immune checkpoints in CGGA and TCGA databases (Figure 5C-5H). The results showed that ALDH enzymes in the same group always had a highly similar immune checkpoint expression status. This suggested that immune functions, particularly checkpoints, may have significant influence over the classification of ALDH function. To explore which cell-types express different subtypes of ALDH enzymes. We analysed different sub-classify ALDH enzyme in distinct cell types based on a public scRNA-seq dataset from 28 patients with GBM (GSE131928).These 24,131 single cells were divided into four cell types including macrophages, malignant cells, oligodendrocytes and T cells. The dot plot showed that a majority of ALDH isoforms, including ALDH16A1, ALDH18A1, ALDH1B1, ALDH1L1/2, ALDH3A2, ALDH4A1, ALDH5A1, ALDH6A1, ALDH7A1, ALDH8A1 and ALDH9A1 were enriched in malignant cells, while ALDH16A1, ALDH2, ALDH3B1 tended to enrich in tumor infiltrating macrophages. ALDH3A2 and ALDH6A1 were enriched in oligodendrocytes. Interestingly, all of the ALDH isoforms expression in tumor infiltrating T cells were lower than those in malignant cells (Figure S5).
ALDH functional classification can be used to improve the efficacy of tumor immunotherapy
As a new star in tumor immunotherapy, immune checkpoint inhibitors have entered clinical practice and shown good clinical benefits in several solid tumors. However, they have not shown an excellent therapeutic effect in the treatment of gliomas. Based on the above results, we wondered if ALDH functional subgroups can improve the immunotherapy effects for glioma treatment. As shown in Figure 6 and Figure S6, in patients with high immune checkpoint gene expression and high expression of ALDH enzymes in Groups I and II, their prognosis was not worse than that of patients with low immune checkpoint gene expression. Furthermore, patients with high immune checkpoint gene expression and low expression of ALDH enzymes in Group IV also had a worse survival rate. This result provided a theoretical basis for the clinical application of specific inhibitors or activators of the corresponding ALDH subgroups, rather than pan-ALDH activity activators or inhibitors.

Discussion
The vital biochemical functions of ALDH in cancer has become a focal point of cancer research. Recent studies have also confirmed ALDH as a novel stem cell marker for various solid tumors, such as glioma, lung cancer, breast cancer, and cholangiocarcinoma (9,10,14-16). There are also many targeted treatments for ALDH in cancers (17). However, no efforts have yet to succeed in improving patient outcomes in the clinic. There may be 2 reasons for this lack of progress: (I) in cancer research, almost all research has focused on enzyme activity or only one specific ALDH isoform. As the ALDH enzyme contains 19 isoforms, some different isoforms are likely to have similar biological functions in cancer. Therefore, it is not surprising that treatments targeting a single isoform were unsuccessful. (II) In the study of isoforms of enzymes, biochemists have classified ALDH isoforms based on their protein structure or substrates in normal cells (18). However, in the unique metabolic environment of tumors, the isoforms of ALDH are likely to exert completely different biological functions to those of normal cells. Therefore, the functional classification of ALDH isoforms in tumors is imperative.
Unlike the methods used by biochemists, we grouped ALDH isoforms based on their biological functions and signaling activations in glioma. This integrated clustering result was mutually verifiable in a large cohort of 999 glioma patients from 2 independent databases. Traditional clusters based on mRNA and methylation cannot be repeated in 2 databases. Our analysis also found that functional and pathway clustering analysis was inherently flawed, which was different from that of traditional clustering analysis. Isoforms in the same subgroup had similar functional characteristics, which were different from those of other subgroups. Although almost all previous studies have shown that ALDH enzymes could promote the malignant progression of tumors, our study surprisingly found that isoforms in some subgroups may inhibit glioma progression. Subsequently, the predictive analysis also confirmed that higher expression of isoforms in subgroups for which glioma was inhibited led to a better prognosis. These results further supported the reliability of the results from the functional analysis. Unsurprisingly, we found that isoforms in different subgroups affected the prognosis of glioma patients by regulating proliferation, transcription, and apoptosis of tumor cells (8,19). Similar results can be seen in previous ALDH related cancer research (16,20,21).
To investigate whether ALDH affected glioma progression by another unexpected function, we explored the relationship between ALDH subgroups and gene mutation counts. Interestingly, we found a close relationship between some subgroups of ALDH and genomic stability. The further in-depth analysis showed that altered DNA repair functions caused by some ALDH subgroups were among the reasons for this phenomenon. However, only a few existing studies have encompassed the relationship between ALDH enzymes and DNA repair functions. One study examined the DNA repair functions of ALDH in alcoholic liver damage, and another analyzed the effects of ALDH1A1 on DNA repair functions in ovarian cancer stem cells (22,23). Our study found a close relationship between ALDH enzymes and DNA repair functions in gliomas and revealed that this mechanism might be an essential cause of the poor prognosis of patients with glioma. This finding provides a new consideration for the use of ALDH functional study in glioma treatment.
We also found that DNA repair functions most relevant to ALDH subgroups were closely related to the immune functions in glioma. Different subgroups had significantly different immune statuses, one of the reasons for which was that ALDH regulated the expression of immune checkpoints. Although few reports have covered the relationship between ALDH and immune functions in other cancers, all studies have regarded ALDH as a stem marker for targeted elimination (24,25). However, our study found that a ALDH enzymes had a robust immune regulation function. We also confirmed that only a small number of glioma patients could benefit from pan-ALDH targeted therapy. However, based on ALDH functional classification, almost all glioma patients could benefit from immunotherapy. This is another innovative discovery of this research.
This study has provided a novel approach for functional analysis of multi-subtype metabolic enzymes in diseases and yielded supporting data for basic and clinical studies of ALDH in glioma. Furthermore, we also revealed several new potential functions and regulatory mechanisms of ALDH in glioma, even providing a possible solution to the dilemma of immunotherapy. However, this functional analysis approach was based on mRNA sequencing data, and more experimental data is needed to valid its accuracy. The clinical application of this research depends on the development of ALDH isoform-specific activators and inhibitors (26-29). Considering ALDH is an essential metabolic enzyme, it also plays an essential role in the average human brain. We were excited to find that different ALDH isoforms have different levels of importance in the metabolism of the average human brain and glioma cells. Therefore, the application of a future ALDH drug cocktail will have better efficacy and fewer side effects. Such an application will enhance the meaningfulness and clinical application of our research.
In conclusion, this study comprehensively provided a novel approach to reveal different biological functions of distinct subtypes of ALDH enzymes in glioma. Our data showed a strong relationship between distinct subtypes of ALDH enzymes, immune regulation functions, and DNA repair function of glioma, which provided a basis for developing relevant immunotherapy strategies.
Acknowledgments
We thank Ms. Shuqing Sun and Hua Huang for tissue sample collection and clinical data retrieval. We thank C. Mullens and J. Gray for modifying the manuscript.
Funding: This research was funded by grants from the National Natural Science Foundation of China (No. 82072768), Construction of the Genomics Platform for Chinese People’s Brain Diseases (No. PXM2019_026280_000002), Sino-German Center Cooperation and Exchanges Program (M-0020), The Medical and Health Technology Innovation Project of the Chinese Academy of Medical Sciences (2020-I2M-C&T-A-024) and National Natural Science Foundation of China (No. 82002994).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tcr-21-1160
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-1160
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1160). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Tiantan Hospital, Capital Medical University (KY2020-093-02). The institutional review board decided that informed consent was not required.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987-96. [Crossref] [PubMed]
- Bouffet E, Larouche V, Campbell BB, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol 2016;34:2206-11. [Crossref] [PubMed]
- Hu H, Mu Q, Bao Z, et al. Mutational Landscape of Secondary Glioblastoma Guides MET-Targeted Trial in Brain Tumor. Cell 2018;175:1665-1678.e18. [Crossref] [PubMed]
- Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765-73. [Crossref] [PubMed]
- Pusch S, Krausert S, Fischer V, et al. Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol 2017;133:629-44. [Crossref] [PubMed]
- Johanns TM, Miller CA, Dorward IG, et al. Immunogenomics of Hypermutated Glioblastoma: A Patient with Germline POLE Deficiency Treated with Checkpoint Blockade Immunotherapy. Cancer Discov 2016;6:1230-6. [Crossref] [PubMed]
- Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007;1:555-67. [Crossref] [PubMed]
- Mao P, Joshi K, Li J, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A 2013;110:8644-9. [Crossref] [PubMed]
- Chen MH, Weng JJ, Cheng CT, et al. ALDH1A3, the Major Aldehyde Dehydrogenase Isoform in Human Cholangiocarcinoma Cells, Affects Prognosis and Gemcitabine Resistance in Cholangiocarcinoma Patients. Clin Cancer Res 2016;22:4225-35. [Crossref] [PubMed]
- Shao C, Sullivan JP, Girard L, et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res 2014;20:4154-66. [Crossref] [PubMed]
- Butowski N, Prados MD, Lamborn KR, et al. A phase II study of concurrent temozolomide and cis-retinoic acid with radiation for adult patients with newly diagnosed supratentorial glioblastoma. Int J Radiat Oncol Biol Phys 2005;61:1454-9. [Crossref] [PubMed]
- Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014;158:929-44. [Crossref] [PubMed]
- Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018;33:676-689.e3. [Crossref] [PubMed]
- Rasper M, Schäfer A, Piontek G, et al. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro Oncol 2010;12:1024-33. [Crossref] [PubMed]
- Xu X, Chai S, Wang P, et al. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett 2015;369:50-7. [Crossref] [PubMed]
- Charafe-Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res 2010;16:45-55. [Crossref] [PubMed]
- Cheng P, Wang J, Waghmare I, et al. FOXD1-ALDH1A3 Signaling Is a Determinant for the Self-Renewal and Tumorigenicity of Mesenchymal Glioma Stem Cells. Cancer Res 2016;76:7219-30. [Crossref] [PubMed]
- Marchitti SA, Deitrich RA, Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol Rev 2007;59:125-50. [Crossref] [PubMed]
- Wu W, Schecker J, Würstle S, et al. Aldehyde dehydrogenase 1A3 (ALDH1A3) is regulated by autophagy in human glioblastoma cells. Cancer Lett 2018;417:112-23. [Crossref] [PubMed]
- Yang L, Ren Y, Yu X, et al. ALDH1A1 defines invasive cancer stem-like cells and predicts poor prognosis in patients with esophageal squamous cell carcinoma. Mod Pathol 2014;27:775-83. [Crossref] [PubMed]
- Pérez-Alea M, McGrail K, Sánchez-Redondo S, et al. ALDH1A3 is epigenetically regulated during melanocyte transformation and is a target for melanoma treatment. Oncogene 2017;36:5695-708. [Crossref] [PubMed]
- Smith C, Gasparetto M, Jordan C, et al. The effects of alcohol and aldehyde dehydrogenases on disorders of hematopoiesis. Adv Exp Med Biol 2015;815:349-59. [Crossref] [PubMed]
- Meng E, Mitra A, Tripathi K, et al. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS One 2014;9:e107142. [Crossref] [PubMed]
- Bazewicz CG, Dinavahi SS, Schell TD, et al. Aldehyde dehydrogenase in regulatory T-cell development, immunity and cancer. Immunology 2019;156:47-55. [Crossref] [PubMed]
- Maccalli C, Parmiani G, Ferrone S. Immunomodulating and Immunoresistance Properties of Cancer-Initiating Cells: Implications for the Clinical Success of Immunotherapy. Immunol Invest 2017;46:221-38. [Crossref] [PubMed]
- Morgan CA, Parajuli B, Buchman CD, et al. N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem Biol Interact 2015;234:18-28. [Crossref] [PubMed]
- Yang SM, Yasgar A, Miller B, et al. Discovery of NCT-501, a Potent and Selective Theophylline-Based Inhibitor of Aldehyde Dehydrogenase 1A1 (ALDH1A1). J Med Chem 2015;58:5967-78. [Crossref] [PubMed]
- Canuto RA, Muzio G, Salvo RA, et al. The effect of a novel irreversible inhibitor of aldehyde dehydrogenases 1 and 3 on tumour cell growth and death. Chem Biol Interact 2001;130-132:209-18. [Crossref] [PubMed]
- Thomas ML, de Antueno R, Coyle KM, et al. Citral reduces breast tumor growth by inhibiting the cancer stem cell marker ALDH1A3. Mol Oncol 2016;10:1485-96. [Crossref] [PubMed]


