
Carfilzomib inhibits the proliferation and apoptosis of multiple myeloma cells by inhibiting STAT1/COX-2/iNOS signaling pathway
Introduction
Cancer is a major global health problem, causing nearly 7.6 million deaths worldwide, and it is estimated that the number of cancers will reach 21 million by 2030. In addition to external factors such as smoking, alcohol, and radiation, there are also internal factors such hormone imbalance, gene mutations and disorders of the human immune system, which can cause abnormal proliferation of normal cells and their transformation into malignant cells (1). For example, multiple myeloma (MM) is a common hematologic malignancy caused by clonal expansion of malignant plasma cells.
Although there are many methods of treatments, including high-dose chemotherapy, stem cell transplantation, and combinations of some innovative compounds [e.g., proteasome inhibitors (PIs), immunomodulators, and antibodies] or above, they remain incurable in most cases despite the introduction and approval of multiple new drugs (2-5). Therefore, new and more effective approaches are urgently needed, especially in the case of relapsed or refractory disease. The ubiquitin-proteasome pathway (UPP) plays a key role in the recognition and degradation of abnormal and misfolded proteins (6). The protein is degraded by the proteasome by covalent attachment of 8.5 kDa ubiquitin, and ubiquitination occurs with the assistance of three different enzymes. In the protein degradation pathway, the 26S proteasome is the major proteolytic component, containing two ATP-dependent 19S regulatory particles and a 20S core particle. The 20S proteasome is a key element of the protease activity of UPP, which dominates most of the intracellular protein degradation in eukaryotic cells. Proteasome-targeted drugs have been demonstrated to play an important role in the treatment of diseases such as MM (7,8). Carfilzomib is a selective PI that irreversibly binds the proteasome (9,10). It exerts its antimyeloma activity through a variety of effects, including induction of undeployed protein stress response (11), downregulation of nuclear factor kappa B (NF-κB) pro-survival activity (12), and changes in bone turnover and the bone marrow microenvironment leading to increased bone strength and diminished microenvironment support of myeloma cells (13-15). Immunogenic MM cell death is induced by increased natural killer cell-mediated lysis of MM cells and enhanced antigen presentation (16-19). Signal transducer and activator of transcription 1 (STAT1) is activated by a variety of cytokines, such as interferons (IFNs), tumor necrosis factor (TNF)-α, interleukin (IL)-6, Platelet derived growth factor (PDGF), and Epidermal Growth Factor (EGF) (20). Phosphorylation of STAT1 by the Janus kinase (JAK) family of kinases results in homology and heterodimerization. After being transferred to the nucleus, STAT1 participates in a variety of cellular processes, such as proliferation, differentiation and cell death (21). According to most studies, STAT1 is considered to be a tumor suppressor, for example in melanoma and ovarian cancer, lung cancer, colon cancer, esophageal cancer, head and neck cancer, liver cancer, and gastric cancer (22-25). On the other hand, some studies support the concept of STAT1 as a tumor promoter, for example in breast cancer, sarcoma, Wilms' tumor, and leukemia (26,27). STAT1 induces tumor cell proliferation by upregulating various proinflammatory factors and chemokines such as TNF-α, cyclooxygenase (COX)-2 and inducible nitric oxide synthase (iNOS) (28). Activation of STAT1 also stimulates the expression of anti-apoptotic genes, such as inhibitor of apoptosis 1 and 2, Bcl-2 protein family, caspase family and Tumor necrosis receptor-associated factor (TRAF1), being involved in anti-apoptotic cell death of Hodgkin’s lymphoma cells (29). Currently, the USFDA has approved three PIs (bortezomib, cafezomib, and isazomib) for the treatment of MM. In addition, covalent binding with these groups in a variety of short peptides and the false peptide has been explored for cancer treatment. The reactive electrophilic group, however, leads to overreaction, low specificity and instability of these inhibitors and the stability of the covalent interaction is irreversible, which is considered during the treatment as the main cause of side effects (resistance) (30). Overcoming chemoresistance is arguably the most important challenge in cancer therapy today. MM cells may exhibit a variety of constitutive or acquired resistance mechanisms, some of which may even be promoted by chemotherapy treatment. For example, upregulation of the binding of MM cells to fibronectin by multidrug resistance genes or p-glycoprotein will result in cell adhesion-mediated resistance. A variety of cytokines such insulin-like IGF-1, IL-6, vascular endothelial growth factor and TNF-α can trigger signaling pathways, support MM cell proliferation and reduce apoptosis (31). Most of these inhibitors fail to treat solid cancers, mainly due to the rapid covalent binding of proteasomes, thus limiting their widespread distribution in tissues. Therefore, in the present study we explored the potential molecular mechanisms of PIs such as carfilzomib as therapeutic agents in MM.
We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2534/rc).
Methods
Cell culture and reagents
Human MM cell lines MOLP-8, RPMI-8226, NCI-H929 and OPM-2 (Cell source: Shanghai Cell Bank, Chinese Academy of Sciences) were cultured at 37 °C in a humidified 5% CO2 incubator with RPMI1640 medium supplemented with 10–20% fetal calf serum (FCS), 100 U/mL penicillin and 100 µg/mL streptomycin (PAA Laboratories, Pasching, Austria). All cell cultures were routinely checked for absence of mycoplasma infection. Carfilzomib stock solution of 100 µM was prepared with 5 mM trifluoroacetic acid dimethyl sulfoxide solution (5 mM trifluoroacetic acid dimethyl sulfoxide solution) and stored at −20 °C. The MM cell lines (MOLP-8, RPMI-8226, NCI-H929, and OPM-2) were exposed to different (1, 5, 10, 15, 20, and 25 nM) concentrations of carfilzomib for 48 h.
Cell extract preparation and western blot analysis
Cultured human MM cells were washed twice with phosphate-buffered saline (PBS) (PAA Laboratories, Pasching, Austria), using RIPA cracking liquid (100 µL, 1 M Tris, pH 7.0. 200 µL, 10% SDS; 100 µL β mercaptoethanol; 1,314.3 µL ddH2O with 285.7 µL complete protease inhibitor [Roche Diagnostics, Indianapolis, IN, USA)] for total protein extraction. Protein quantification was performed by the BCA method, and 5× protein loading buffer was added to the supernatant at 1:4, and denatured by heating in boiling water for 10 min. Next, 50 µg of protein was subjected to polyacrylamide gel (SDS-PAGE) electrophoresis for protein separation, the separated protein was transferred to PVDF membrane by wet transfer method, and the PVDF membrane was sealed with 5% skim milk at room temperature for 2 hours. Caspase3 (1:1,000), Bax (1:1,000), Bcl2 (1:1,000), STAT1 (1:1,000), COX2 (1:1,000), iNOS (1:1,000) and β-actin (1:2,000) primary antibodies (Cell Signaling Technology, Danvers, Massachusetts, USA) were added, and incubated overnight at 4 °C while being shaken. The TBST solution was washed three times, 5 min/time, incubated with horseradish enzyme-labeled secondary antibody (1:5,000) for 1 h at room temperature, and washed three times, 5 min/time, with TBST solution. Finally, ECL luminescent solution was evenly applied and photographed on a gel imager for exposure. ImageJ software was used to determine the gray values of the bands, and β-actin was used as an internal reference. The above procedures were repeated three times.
Apoptosis detection (DNA fragment analysis)
To detect apoptosis, we use an enzyme-linked immunosorbent assay (ELISA) + apoptosis assay for cell death detection in accordance with the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). Cells were cultivated under (1, 5, 10, 15, 20, 25 nM) Carfilzomib, Cell lysate is used to lysate cells, and with 400 g centrifugation for 10 min. An equal amount of supernatant was added to a 96-well microdrop plate coated with streptavidin. Anti-DNA-POD and anti-histone solutions were added and the plates were incubated on a platform rotator for 2 h at room temperature. The generation of histone-bound DNA fragments showed green and was quantified by spectrophotometer (Thermo Scientific Appliscan, Waltham, MA, USA) at a wavelength of 405 nm. The apoptosis rate of treated cells was calculated as a percentage of the drug loading control.
Apoptosis detection (TUNEL staining)
Twenty-five thousand cells were inoculated into each well of 96-well plates contained 100 microLiters of RPMI and 10–20% FCS, Carfilzomib (5, 10, 20 nM) was treated for 48 h, the culture medium was removed by suction, washed three times with 1× PBS for 5 min/time, fixed with 4% PFA for 30 min at room temperature, washed three times with 1× PBS for 5 min/time, then 100 µL of 20 µg/mL proteinase K solution was added to each well before incubation at room temperature for 20 min, followed by washing three times with 1× PBS for 5 min/time, addition of 100 µL of equilibration solution and further incubation at room temperature for 10 min. The equilibration solution was removed by suction, 50 µL of reaction solution (Roche Diagnostics, Mannheim, Germany) was added, the plates were incubated at 37 °C for 1 h, washed three times with 1× PBS for 5 min/time, then 100 µL of Hoechst was added to each well before incubation at room temperature for 20 min, washing three times with 1× PBS for 5 min/time, and finally observation and imaging under an inverted fluorescence microscope, using Image J software to determine the proportion of TUNEL-positive cells.
Cell survival assay (MTT)
Twenty-five thousand cells were inoculated into each well of 96-well plates contained 100 microLiters of RPMI and 10–20% FCS. At 48 h after treatment, the number of living cells was determined in aqueous solution by cell proliferation test, according to the manufacturers instructions (Promega, Mannheim, Germany). Quantitative detection was performed at a wavelength of 570 nm using a spectrophotometer (Thermo Scientific) and using 630 nm wavelength as the reference minus the background signal.
Cell survival assay (CCK8)
A 96-well plate containing 100 microl RPMI and 10–20% FCS was inoculated with 25,000 cells per well. After 48 h of treatment, cell survival rate was detected by CCK8 kit (GLPBIO, USA). Quantitative detection was performed using a spectrophotometer with wavelength of 450 nm (Thermo Scientific), with wavelength of 630 nm as reference, and background signal was subtracted.
Proliferation assay (5-bromo-2′-deoxyuridine incorporation)
To determine the proliferation of MM cells, 25,000 cells were implanted in a 96-well culture plate. Carfilzomib (5, 10, 20 nM) was treated for 48 h. Thymine analogue 5-bromo-2'-deoxyuridine (BrdU) was added 4 h before the end of the experiment, and cells were collected at the end point of the experiment 48 h. The proliferation rate was determined by ELISA (Roche diagnosis) and 450 nm bispectral spectrophotometer colorimetry.
Statistical analysis
All data in our study are expressed as mean ± standard error (mean ± SEM) and were analyzed using Prism 7.0 software (GraphPad, La Jolla, CA, USA). Behavioral experimental data were analyzed by one-way analysis of variance (ANOVA) with repeated measures, followed by Dunnett’s test. Comparisons were performed by ANOVA followed by Dunnett’s test. P values <0.05 were considered statistically significant.
Results
Structure of carfilzomib
The proteasome inhibitor carfilzomib has been approved by the USFDA for the treatment of MM (Figure 1). It is a covalent peptidyl inhibitor with an electrophilic portion at the c-terminus of the peptidyl backbone that covalently binds to the Thr1 residue at the catalytic end of the β5 subunit of the proteasome.
Effect of carfilzomib on apoptosis and cell viability of MM cell lines
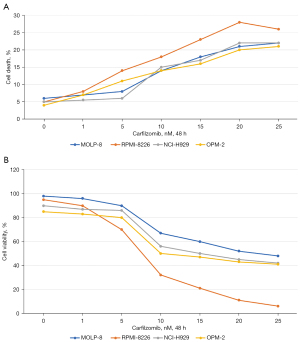
The MM cell lines (MOLP-8, RPMI-8226, NCI-H929, and OPM-2) were exposed to different (1, 5, 10, 15, 20, and 25 nM) concentrations of carfilzomib for 48 h. Cells were detected by flow cytometry after 7-AAD staining, and carfilzomib showed dose-dependent induction of apoptosis in all MM cell lines (Figure 2A). The apoptotic rates after 48 h of incubation were 15.20%±0.2% MOLP-8, 20.73%±0.21% RPMI-8226, 16.55%±2.00% NCI-H929, and 15.00%±2.84% OPM-2 cells. In the MTT assay of viable cells, carfilzomib showed dose-dependent inhibition of cell viability in all MM cell lines (Figure 2B). The IC50 values after 48-h incubation were 12.20±0.14 µM MOLP-8, 10.73±3.21 µM RPMI-8226, 26.15±2.05 µM NCI-H929, and 15.97±1.84 µM for OPM-2 cells. RPMI-8226 cells are more sensitive to carfilzomib than other MM cells. Therefore, that cell line was selected to elucidate the molecular mechanism of carfilzomib cytotoxic activity in more detail.
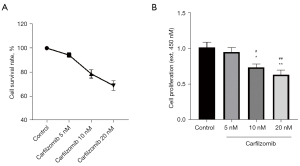
Effect of carfilzomib on proliferation of RPMI-8226 cells
In the MM cells treated with Carfilzomib, the cell survival rate decreased, and the incorporation of BrdU as a proliferation indicator decreased. In addition, cafezomib was dose-dependent on cell survival and proliferation of RMI-8226 cells. The proliferation and survival rate of RPMI-8226 cells decreased by 30% and 28% at 10 nM. At 20 nM, proliferation decreased by 43% and cell viability decreased by 45% (Figure 3).
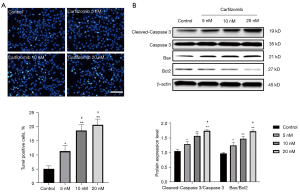
Effect of carfilzomib on apoptosis of RPMI-8226 cells
Upon genomic DNA fragmentation, the exposed 3'-OH can be catalyzed by terminal deoxynucleotidyltransferase (TdT) plus fluorescein (FITC)-labeled dUTP (fluorescein-dUTP), which can be detected by fluorescence microscopy. At present, the Bcl-2 protein family and caspase family attract the most attention, including the Bcl-2 gene and Bax genes, which are known as the most important pair of genes to inhibit apoptosis, and caspase 3 executive protease is the most important in the regulation of apoptosis. When they were used as a parameter to measure apoptosis of RPMI-8226 cells, apoptosis was observed and a significant dose-dependent increase occurred (Figure 4A,4B). In fact, at concentrations >5 nM, the cells all showed a significant apoptotic response (P<0.05 at 5 nM and P<0.01 at 10 and 20 nM). At a concentration of 10 nM, carfilzomib increased the apoptotic rate of RPMI-8226 cells by 3-fold and apoptotic protein expression by 1.5-fold compared with control-treated cells.
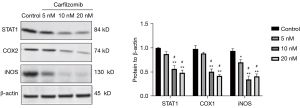
Induction of tumor progression by carfilzomib
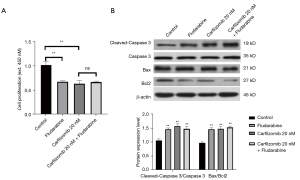
It has been reported that the cytokine TNFα can stimulate the increased expression of STAT1, thus inducing cell stress response and cell proliferation. Therefore, we investigated the STAT1, iNOS and COX-2 protein expression levels in carfilzomib-treated RPMI-8226 cells. All were overexpressed in untreated cells and decreased in a dose-dependent manner after apigenin treatment (Figure 5).
Carfilzomib induces tumor progression through the STAT1/COX2/iNOS signaling pathway
Then Fludarabine (NSC 118218) (Selleck, USA), an inhibitor of STAT1, was introduced for functional reversal. Fludarabine has been reported to inhibit cell proliferation and promote cell apoptosis. RPMI-8226 cells were treated with both Carfilzomib and Fludarabine. The detection results show that the additive has no superposition effect. It was demonstrated that Carfilzomib regulates tumor progression through STAT1 (Figure 6).
Discussion
In 2016, there 138,509 new cases of MM globally, and in America, 30,770 cases are expected to occur in 2018 (32). In recent decades, the global incidence has increased dramatically, in part because of the aging of populations. In MM clinical trials, the introduction of PIs and immunomodulatory drugs has improved treatment responses compared with older therapies for patients with newly diagnosed MM (33). Carfilzomib-based regimens have shown strong efficacy, and carfilzomib has been approved for the treatment of recurrent or refractory MM (34).
The main mechanism of anticancer drugs is apoptosis induction, and multiple signaling pathways are involved in this process (35). In the present study, TUNEL staining showed that carfilzomib increased the proportion of RPMI-8226 cells undergoing apoptosis in a dose-dependent manner. Shrinkage, fragmentation, chromatin condensation, and DNA fragmentation of the cells after treatment were distinct features of apoptosis (35). Treatment of tumor cells with carfilzomib activates the levels of the caspase and Bcl2 families and nuclear condensation, thereby inducing apoptosis. Anticancer compounds usually inhibit cell proliferation by promoting cell cycle arrest (36). Carfilzomib inhibits the DNA synthesis phase (S phase) of MM cells and arrests most of the cells in G2/M phase in a dose-dependent manner (37). In contrast, cell cycle arrest was induced at the G2/M checkpoint in the oral squamous cell carcinoma cell line SCC-25, which inhibited the expression of cyclinD1 and E and the inactivation of CDK1 (38).
It is estimated that more than 20% of human cancers are caused by chronic inflammation. STAT1 is involved in tumorigenesis and metastasis by upregulating inducible enzymes such as COX-2 and iNOS (38-40), which lead to the overproduction of nitric oxide and prostaglandin E2 (41). We examined the expression of STAT1, iNOS and COX-2 in cells after carfilzomib treatment. Carfilzomib inhibited the activation of STAT1, which is responsible for the expression of iNOS and COX-2 in cancer cells. Carfilzomib dose-dependently decreased iNOS and COX-2 expressions compared with untreated cells, which was consistent with other reports showing inhibition of the expressions of iNOS and COX-2 (42-45). Inhibiting the overproduction of nitric oxide and prostaglandin E2 is an effective chemopreventive strategy (46). Inhibition of MAP3K8 may interrupt important interactions between macrophages and cancer cells, leading to dose-dependent apoptosis of MM cells (47). To demonstrate the activity of carfilzomib in hematological tumor cells, we investigated the possible mechanism by which it caused reduced cell survival and increased apoptosis in MM cell lines. Carfilzomib effectively reduced cell viability and induced apoptosis in a dose-dependent manner in all of the studied MM cell lines, with the strongest sensitivity to RPMI-8226 cells. Some studies have reported the expression of STAT1 in RPMI-8226, NCI-H929, KMS-11, OPM-2, AMO-I, l-363, JJN-3, U266, MM.1S, KMS-20BM, and KMS-28BM MM cell lines (48-50). The prognostic role of related enzymes COX-2 and iNOS in MM and other malignancies has been reported (51). Other authors also discuss the involvement of STAT1, COX-2 and iNOS in all types of inflammation, angiogenesis, and metastasis of human cancer, chemical resistance and their special role in the development of drug resistance (52). Our results are consistent with several previous reports, confirming that the STAT1/COX2/iNOS signaling pathway plays a key role in MM (pathology) biology. We then treated the cells with STAT1 inhibitor Fludarabine and Carfilzomib, and found that Fludarabine reversed the effects of Carfilzomib on the proliferation and apoptosis of RPMI-8226 cells. It was further demonstrated that Carfilzomib acted through the STAT1/COX2/iNOS signaling pathway.
To sum up, this study provides strong circumstantial evidence that the new PI inhibitor, carfilzomib, can inhibit growth and induce apoptosis in vitro in MM cells (53). One concern is direct activity of the MM cells themselves. In human patients, however, with complex “natural” environments, and other anatomic sites of MM, multiple interactions of bone marrow cells may not only activate the STAT1/COX2/iNOS pathway, but also activate PI3K/Akt, MEK/ERK, JAK/STAT3, NF-kappa B or other signal cascades that may promote the survival, drug resistance and/or migration of tumor cells (54,55). Therefore, although the efficacy of carfilzomib remains to be further studied, such as in combination with targeted therapy and chemotherapy, our data provides experimental evidence for future research and suggest a path worth exploring.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2534/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2534/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2534/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Matsuo K, Palmer JB. Anatomy and physiology of feeding and swallowing: normal and abnormal. Phys Med Rehabil Clin N Am 2008;19:691-707. vii. [Crossref] [PubMed]
- Nilner M, Lassing SA. Prevalence of functional disturbances and diseases of the stomatognathic system in 7-14 year olds. Swed Dent J 1981;5:173-87. [PubMed]
- Weinberg LA. The biomechanics of force distribution in implant-supported prostheses. Int J Oral Maxillofac Implants 1993;8:19-31. [PubMed]
- Ramos Verri F, Santiago JF Junior, de Faria Almeida DA, et al. Biomechanical influence of crown-to-implant ratio on stress distribution over internal hexagon short implant: 3-D finite element analysis with statistical test. J Biomech 2015;48:138-45. [Crossref] [PubMed]
- Buschang PH, Hayasaki H, Throckmorton GS. Quantification of human chewing-cycle kinematics. Arch Oral Biol 2000;45:461-74. [Crossref] [PubMed]
- Eskitascioglu G, Usumez A, Sevimay M, et al. The influence of occlusal loading location on stresses transferred to implant-supported prostheses and supporting bone: A three-dimensional finite element study. J Prosthet Dent 2004;91:144-50. [Crossref] [PubMed]
- Takahashi JM, Dayrell AC, Consani RL, et al. Stress evaluation of implant-abutment connections under different loading conditions: a 3D finite element study. J Oral Implantol 2015;41:133-7. [Crossref] [PubMed]
- Steinebrunner L, Wolfart S, Ludwig K, et al. Implant-abutment interface design affects fatigue and fracture strength of implants. Clin Oral Implants Res 2008;19:1276-84. [Crossref] [PubMed]
- Quek HC, Tan KB, Nicholls JI. Load fatigue performance of four implant-abutment interface designs: effect of torque level and implant system. Int J Oral Maxillofac Implants 2008;23:253-62. [PubMed]
- Wang RF, Kang B, Lang LA, et al. The dynamic natures of implant loading. J Prosthet Dent 2009;101:359-71. [Crossref] [PubMed]
- Alkan I, Sertgöz A, Ekici B. Influence of occlusal forces on stress distribution in preloaded dental implant screws. J Prosthet Dent 2004;91:319-25. [Crossref] [PubMed]
- Zipprich H, Rathe F, Pinz S, et al. Effects of Screw Configuration on the Preload Force of Implant-Abutment Screws. Int J Oral Maxillofac Implants 2018;33:e25-32. [Crossref] [PubMed]
- Uhl RL. The biomechanics of screws. Orthop Rev 1989;18:1302-7. [PubMed]
- Özkurt Z, Kazazoğlu E. Zirconia dental implants: a literature review. J Oral Implantol 2011;37:367-76. [Crossref] [PubMed]
- Andreiotelli M, Wenz HJ, Kohal RJ. Are ceramic implants a viable alternative to titanium implants? A systematic literature review. Clin Oral Implants Res 2009;20:32-47. [Crossref] [PubMed]
- Johansson CB, Han CH, Wennerberg A, et al. A quantitative comparison of machined commercially pure titanium and titanium-aluminum-vanadium implants in rabbit bone. Int J Oral Maxillofac Implants 1998;13:315-21. [PubMed]
- Cinel S, Celik E, Sagirkaya E, et al. Experimental evaluation of stress distribution with narrow diameter implants: A finite element analysis. J Prosthet Dent 2018;119:417-25. [Crossref] [PubMed]
- Park JH, Lim YJ, Kim MJ, et al. The effect of various thread designs on the initial stability of taper implants. J Adv Prosthodont 2009;1:19-25. [Crossref] [PubMed]
- Abuhussein H, Pagni G, Rebaudi A, et al. The effect of thread pattern upon implant osseointegration. Clin Oral Implants Res 2010;21:129-36. [Crossref] [PubMed]
- O'Sullivan D, Sennerby L, Jagger D, et al. A comparison of two methods of enhancing implant primary stability. Clin Implant Dent Relat Res 2004;6:48-57. [Crossref] [PubMed]
- Raaj G, Manimaran P, Kumar CD, et al. Comparative Evaluation of Implant Designs: Influence of Diameter, Length, and Taper on Stress and Strain in the Mandibular Segment-A Three-Dimensional Finite Element Analysis. J Pharm Bioallied Sci 2019;11:S347-54. [Crossref] [PubMed]
- Lee JH, Huh YH, Park CJ, et al. Effect of the Coronal Wall Thickness of Dental Implants on the Screw Joint Stability in the Internal Implant-Abutment Connection. Int J Oral Maxillofac Implants 2016;31:1058-65. [Crossref] [PubMed]
- Marcián P, Wolff J, Horáčková L, et al. Micro finite element analysis of dental implants under different loading conditions. Comput Biol Med 2018;96:157-65. [Crossref] [PubMed]
- Chun HJ, Cheong SY, Han JH, et al. Evaluation of design parameters of osseointegrated dental implants using finite element analysis. J Oral Rehabil 2002;29:565-74. [Crossref] [PubMed]
- Cehreli M, Duyck J, De Cooman M, et al. Implant design and interface force transfer. A photoelastic and strain-gauge analysis. Clin Oral Implants Res 2004;15:249-57. [Crossref] [PubMed]
- Pico A, Martín-Lancharro P, Caneiro L, et al. Influence of abutment height and implant depth position on interproximal peri-implant bone in sites with thin mucosa: A 1-year randomized clinical trial. Clin Oral Implants Res 2019;30:595-602. [Crossref] [PubMed]
- Pommer B, Bucur L, Zauza K, et al. Meta-analysis of oral implant fracture incidence and related determinants. J Oral Implants 2014;2014:7. [Crossref]
- Miyamoto I, Tsuboi Y, Wada E, et al. Influence of cortical bone thickness and implant length on implant stability at the time of surgery--clinical, prospective, biomechanical, and imaging study. Bone 2005;37:776-80. [Crossref] [PubMed]
- Buser D, Sennerby L, De Bruyn H. Modern implant dentistry based on osseointegration: 50 years of progress, current trends and open questions. Periodontol 2000 2017;73:7-21. [Crossref] [PubMed]
- Cooper LF. Biologic determinants of bone formation for osseointegration: clues for future clinical improvements. J Prosthet Dent 1998;80:439-49. [Crossref] [PubMed]
- Delgado-Ruiz RA, Calvo-Guirado JL, Romanos GE. Effects of occlusal forces on the peri-implant-bone interface stability. Periodontol 2000 2019;81:179-93. [Crossref] [PubMed]
- Szmukler-Moncler S, Salama H, Reingewirtz Y, et al. Timing of loading and effect of micromotion on bone-dental implant interface: review of experimental literature. J Biomed Mater Res 1998;43:192-203. [Crossref] [PubMed]
- Frost HM. Strain and other mechanical influences on bone strength and maintenance. Curr Opin Orthop 1997;8:60-70. [Crossref]
- Frost HM. Bone's mechanostat: a 2003 update. Anat Rec A Discov Mol Cell Evol Biol 2003;275:1081-101. [Crossref] [PubMed]
- Skerry TM. One mechanostat or many? Modifications of the site-specific response of bone to mechanical loading by nature and nurture. J Musculoskelet Neuronal Interact 2006;6:122-7. [PubMed]
- Kamijou T, Nakajima T, Ozawa H. Effects of osteocytes on osteoinduction in the autogenous rib graft in the rat mandible. Bone 1994;15:629-37. [Crossref] [PubMed]
- Hughes JM, Petit MA. Biological underpinnings of Frost's mechanostat thresholds: the important role of osteocytes. J Musculoskelet Neuronal Interact 2010;10:128-35. [PubMed]
- Frost HM. From Wolff's law to the Utah paradigm: insights about bone physiology and its clinical applications. Anat Rec 2001;262:398-419. [Crossref] [PubMed]
- Ashman RB, Van Buskirk WC. The elastic properties of a human mandible. Adv Dent Res 1987;1:64-7. [Crossref] [PubMed]
- Martin AD, McCulloch RG. Bone dynamics: stress, strain and fracture. J Sports Sci 1987;5:155-63. [Crossref] [PubMed]
- Aboushelib MN. Simulation of cumulative damage associated with long term cyclic loading using a multi-level strain accommodating loading protocol. Dent Mater 2013;29:252-8. [Crossref] [PubMed]
- Schaffler MB, Radin EL, Burr DB. Mechanical and morphological effects of strain rate on fatigue of compact bone. Bone 1989;10:207-14. [Crossref] [PubMed]
- Carter DR, Caler WE, Spengler DM, et al. Fatigue behavior of adult cortical bone: the influence of mean strain and strain range. Acta Orthop Scand 1981;52:481-90. [Crossref] [PubMed]
- Maminskas J, Puisys A, Kuoppala R, et al. The Prosthetic Influence and Biomechanics on Peri-Implant Strain: a Systematic Literature Review of Finite Element Studies. J Oral Maxillofac Res 2016;7:e4. [Crossref] [PubMed]
- Torres AM, Trikanad AA, Aubin CA, et al. Bone-inspired microarchitectures achieve enhanced fatigue life. Proc Natl Acad Sci U S A 2019;116:24457-62. [Crossref] [PubMed]
- Lin D, Li Q, Li W, et al. Mandibular bone remodeling induced by dental implant. J Biomech 2010;43:287-93. [Crossref] [PubMed]
- I-Chiang C. Finite element modelling of implant designs and cortical bone thickness on stress distribution in maxillary type IV bone. Comput Methods Biomech Biomed Engin 2014;17:516-26. [Crossref] [PubMed]
- Merheb J, Van Assche N, Coucke W, et al. Relationship between cortical bone thickness or computerized tomography-derived bone density values and implant stability. Clin Oral Implants Res 2010;21:612-7. [Crossref] [PubMed]
- Rozé J, Babu S, Saffarzadeh A, et al. Correlating implant stability to bone structure. Clin Oral Implants Res 2009;20:1140-5. [Crossref] [PubMed]
- Stanford CM, Brand RA. Toward an understanding of implant occlusion and strain adaptive bone modeling and remodeling. J Prosthet Dent 1999;81:553-61. [Crossref] [PubMed]
- Unsal GS. Three-Dimensional Finite Element Analysis of Different Implant Configurations in Enlarged First Molar Areas. Int J Oral Maxillofac Implants 2020;35:675-83. [Crossref] [PubMed]
- Joda T, Zarone F, Ferrari M. The complete digital workflow in fixed prosthodontics: a systematic review. BMC Oral Health 2017;17:124. [Crossref] [PubMed]
- Rupp F, Liang L, Geis-Gerstorfer J, et al. Surface characteristics of dental implants: A review. Dent Mater 2018;34:40-57. [Crossref] [PubMed]
- Park S, Kim H, Choi KS, et al. Graphine-Chitosan hybrid dental implants with enhanced antibacterial and cell proliferation properties. Appl Sci 2020;10:4888. [Crossref]
- Lucaciu O, Soriţău O, Gheban D, et al. Dental follicle stem cells in bone regeneration on titanium implants. BMC Biotechnol 2015;15:114. [Crossref] [PubMed]



