
Knockdown of TRIM44 inhibits the progression of ovarian cancer and is related to the FOXM1-EZH2 signaling pathway
Introduction
Ovarian cancer (OC) is one of the most common human malignancies in women, resulting in an estimated 239,000 new diagnoses worldwide each year (1). Epithelial ovarian cancer (EOC) is the most common histological subtype of OC, accounting for 80% to 90% of cases. Approximately 70% of EOC patients are diagnosed with advanced-stage disease, which has the highest mortality rate among all female malignancies. In China, there were approximately 52,100 new cases and 22,500 deaths attributed to EOC in 2015 (2). The prognosis for patients suffering from EOC remains poor due to limited therapeutic strategies and late diagnosis. Despite improved treatments for OC, including debulking surgery, platinum-based chemotherapy and targeted therapy, the 5-year survival rate of advanced OC is only approximately 30% (3). Therefore, investigating predicative biomarkers and understanding the molecular mechanisms underlying EOC development are important for developing effective therapeutic targets.
Tripartite motif-containing protein 44 (TRIM44), a member of the TRIM family, has been documented to be overexpressed in various types of cancer, such as osteosarcoma (4), lung cancer (5), breast cancer (6)], papillary thyroid cancer (7), testicular germ cell tumor (8) and prostate cancer (9). In addition, high expression of TRIM44 is significantly associated with poor prognosis in malignant tumors (10,11). Our previous study demonstrated that high expression of TRIM44 protein in ovarian tissues is significantly correlated with clinicopathological factors and poor prognosis (12). It was all reported that TRIM44 facilitated OC proliferation, migration, and invasion by inhibiting FRK (13). However, expression of TRIM44 in ovarian cell lines and its mRNA levels in ovarian tissues still need to be explored. In addition, the biological functions and carcinogenic mechanisms of TRIM44 in EOC remain unclear.
Here, we report that TRIM44 protein and mRNA levels are frequently increased in EOC clinical tissues and cell lines. Using lentivirus-driven shRNA knockdown, cell proliferation, migration, invasion, colony formation, and apoptosis analyses and in vivo animal experiments, we demonstrated that TRIM44 promotes EOC cell proliferation, migration, invasion, colony formation and xenograft tumor growth. Furthermore, we showed that TRIM44downregulation inhibits the carcinogenesis of EOC cells in part through suppression of the FOXM1-EZH2 signaling pathway. Our data suggest that TRIM44 may be a potential therapeutic target for the treatment of OC.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2915/rc).
Methods
Patient samples
Fresh EOC samples and noncancerous ovarian tissue samples were collected from Harbin Medical University Cancer Hospital (Harbin, China). All patients underwent maximal cytoreduction followed by platinum-based combination chemotherapy. None of the patients received chemotherapy, radiotherapy or immunotherapy before surgery. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics board of Harbin Medical University Cancer Hospital (No. KY2021-48) and informed consent was taken from all the patients.
Cell lines and cell culture
The human EOC cell lines A2780 and SKOV3 were obtained from the Cell Resource Center of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) containing 10% fetal calf serum (FBS, HyClone, Australia). All cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C.
Lentiviral vector encoding shRNA targeting TRIM44
ShRNA was designed using Ambion Company’s online design tool. The lentiviral vector encoding TRIM44 shRNA was synthesized and packaged by Ruisai Biotechnology Co., Ltd. (Shanghai, China). Lentiviral vectors carrying three types of TRIM44-shRNA was used to screen for maximum silencing efficiency. We electrically transfected the TRIM44-shRNA vectors into SKOV3 and A2780 cells to construct a lentiviral vector encoding shRNA targeting TRIM44, and the empty vector was used as the control. Short hairpin oligonucleotides (shTRIM44: 5'-TGCTGTGAGTCATCAACCTATCCATGGTTTTGGCCACTGACTGACCATGGATATTGATGACTCA-3' and shCon: 5'-CCTGTGAGTCATCAATATCCATGGTCAGTCAGTGGCCAAAACCATGGATAGGTTGATGACTCAC -3') were synthesized and cloned into the lentivirus-based vector pLenti6.3-MCS/V5 DEST (Thermo Electron Corporation, Waltham, MA, USA). Lentivirus production was performed by following a standard protocol involving transfection of 293T cells with the recombinant shRNA vector, together with psPAX2 and pVSVG (Thermo Electron Corporation, Waltham, MA, USA).
Lentivirus infection
SKOV3 and A2780 cells were cultured in six-well plates, and lentiviral vectors with the TRIM44 shRNA sequence or NC shRNA sequence were added at a multiplicity of infection (MOI) of 100 to the plate when the cells reached 70% confluency. The medium was replaced after 48 h of infection, and transfection efficiency was evaluated using quantitative real-time polymerase chain reaction (qRT-PCR) and western blot assays.
Western blot analysis and antibodies
Protein was extracted from tissues and cells for analysis. The samples were boiled at 100 °C for 5 min. Equal amounts of protein were separated by SDS-PAGE followed by transfer to PVDF membranes (Millipore, USA). The membranes were incubated with primary antibodies overnight at 4 °C. The following primary antibodies: mouse anti-human TRIM44 (ab236422; 1:1,000 dilution; Abcam, Cambridge, UK), rabbit anti-human CCND3 (ab112034, 1:1,000 dilution; Abcam, Cambridge, UK), rabbit anti‐human FOXM1 (ab245309, 1:1,000 dilution, Abcam, Cambridge, UK), rabbit anti-human EZH2 (#4905,1:100 dilution, Cell Signaling Technology), rabbit anti-human CCNE2 (ab226388, 1:1,000 dilution; Abcam, Cambridge, UK), rabbit anti-human BIRC5 (PAB18224, 1:1,000 dilution; Abnova, China), and mouse anti-GADPH (AG019, 1:1,000 dilution, Beyotime) as internal controls. Immunoreactive protein bands were detected using an ECL detection system (Cell Signaling Technology, USA).
qRT-PCR
mRNA expression was quantified using real-time RT-PCR. Briefly, total RNA was extracted from the cell lines using TRIzol reagent (Ruisai Biotechnology Co., Ltd., Shanghai). After quantification, a total of 10 ng RNA was used for cDNA synthesis, followed by real-time PCR using the One Step SYBR® PrimeScriptTM RT-PCR Kit II (Ruisai Biotechnology Co., Ltd., Shanghai) according to the manufacturer’s instructions. ACTB was employed as an internal control. The primer sequences were as follows: TRIM44-forward: 5'-GGCACTCTTCCAGCCTTCC-3'; TRIM44-reverse: 5'-GAGCCGCCGATCCACAC-3'; ACTB-forward: 5'-GCTGGTAACTTGTTGTGCGG-3'; ACTB-reverse: 5'-GAGCCGCCGATCCACAC-3'. PCR was carried out for 40 cycles (95 °C for 15 s, 60 °C for 20 s and 72 °C for 20 s) according to the instructions. Relative quantification of TRIM44 mRNA was conducted using the 2-ΔΔCT method, followed by normalization using the internal control.
Cell counting kit-8 (CCK-8) assay
An A CCK-8 assay (CK04, Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was performed to examine cell proliferation. After plating in 96-well plates at an initial density of 5×103 cells/well, SKOV3 and A2780 cells were transfected with shTRIM44 or shNC (lentivirus-infected cells). At each time point, cells in each well were incubated with 10 µL of CCK-8 dye in 90 µL of culture medium for 2 h at 37 °C. Then, the OD values at 450 nm were measured using a VERS Amax Microplate Reader (Molecular Devices Corp, Sunnyvale, CA, USA).
Flow cytometry analysis of apoptosis
SKOV3 and A2780 cells were transfected with TRIM44-shRNA for 48 h. The cells were subsequently harvested by centrifugation at 10,000 × g for 5 min. The cells were washed twice with cold PBS, centrifuged at 10,000 × g for 5 min, and then resuspended in PBS. The cells were resuspended and stained with propidium iodide (PI, 1:100 dilution) and Annexin V-APC (Cell Signaling Technology, Danvers, USA) at 20 °C for 30 min in the dark. The stained cells were analyzed using a BD FACSAria Cell Sorter.
Migration and invasion assays
The invasion and migration assays were performed using Transwell inserts (Corning Incorporated, Corning, NY, USA) with (invasion assay) or without Matrigel (migration assay). For the invasion assay, the Transwell chambers were pretreated with Matrigel (BD Bioscience, Mountain View, CA, USA). First, transfected ovarian cells were suspended in medium without FBS and seeded into the upper chamber. The lower chamber contained RPMI-1640 with10% FBS. After culturing for 48 h at 37 °C, noninvasive cells on the upper chambers were removed using cotton swabs, while the invasive cells on the lower chamber were subsequently fixed and stained with 0.1% crystal violet for 15 min. An inverted microscope (Olympus Corporation, Tokyo, Japan) was used to evaluate invasion and migration. The number of cells in five fields of view was counted under a microscope at ×100 magnification. The cross divides the imaging area, and the number of cells in one area is used to replace the number of cells in a group under the premise that the number of cells is as uniform as possible and there is no excessive number of cells.
Colony formation assay
Following lentiviral infection, SKOV3 and A2780 cells were plated in 6-well plates at a density of 1.5×103 cells/well. Then, the cells were cultured in a humidified incubator at 37 °C with 5% CO2. After 7 days, natural colonies formed, and the cells were washed three times with PBS, fixed in 1.5 mL of 4% paraformaldehyde and stained with crystal violet (Sigma-Aldrich) for 20 min. After the samples were washed twice with PBS, the staining solution was removed, and the cells were air dried. Colony formation was assessed using a light/fluorescence microscope to quantify the colony numbers.
Nude mouse in vivo xenograft assays
Experiments were performed under a project license (No. KY2021-48) granted by the ethics board of Harbin Medical University Cancer Hospital, in compliance with the “guidelines and suggestions laboratory animals” (Ministry of Science and Technology of the People’s Republic of China). A protocol was prepared before the study without registration. Female athymic BALB/c nude mice (aged six weeks, weighing 20–22 g) were obtained from Sipple B&K Laboratory Animal Co., Ltd. (Shanghai, China). Mice were housed in a pathogen-free animal facility and randomly assigned to the control or experimental group (six mice per group). To explore the effects of TRIM44 on tumorigenesis in vivo, cells were intraperitoneal injected subcutaneously into the nude mice. Two groups of nude mice were injected with 50 µL of 108 pfu/mL NC and KD virus on day 0, day 3, day 7, day 10, and day 14. A single mouse was considered an experimental unit. Tumor volume was estimated every three days using the following formula: 0.5 × length × width2. Tumor sizes were measured at least once or twice a week to monitor tumor growth. All mice were euthanized after 23 days, and confounding factors such as the order of treatments and measurements or animal location were recorded. If the volume of the tumor decreased after inoculation and no longer increased, the sample was excluded.
Statistical analysis
Data are expressed as the mean ± SD. Statistical analysis was performed using GraphPad Prism 7.0 software. Differences among groups were analyzed using ANOVA or Student’s t test. Data with P<0.05 were considered statistically significant.
Results
TRIM44 expression is increased in EOC tissues and cell lines
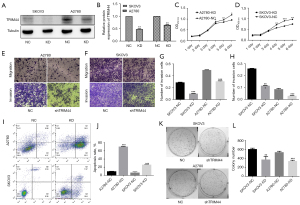
We first examined the mRNA and protein levels of TRIM44 in fresh EOC samples and noncancerous ovarian tissue samples. The results showed that both mRNA and protein expression of TRIM44 were significantly upregulated in EOC samples compared to noncancerous tissue samples (Figure 1; P<0.05), suggesting that TRIM44 may represent a vital biomarker of EOC.

Lentiviral-mediated RNAi significantly inhibits TRIM44 expression in SKOV3 and A2780 cells
The individual shRNA oligos targeting TRIM44 were validated. Western blot results indicated that lentiviral vectors with the TRIM44 shRNA sequence effectively decreased the expression of TRIM44 in SKOV3 and A2780 cells (Figure 2A,2B).
Knockdown of TRIM44 suppresses SKOV3 and A2780 cell proliferation in vitro
To determine whether TRIM44 influenced the malignant properties of EOC cells, we first performed cell proliferation analysis using CCK-8 assays. We found that knockdown of TRIM44 resulted in a significant reduction in cell proliferation (Figure 2C,2D). As shown in Figure 2, the growth curve of shTRIM44-treated cells began to drop on the third day compared to that of the nontreated and shNC-treated cells. The decrease reached 54.14% and 76.93% for shTRIM44-treated SKOV3 and A2780 cells, respectively, on the fifth day compared to shNC-treated cells (P<0.001).
Knockdown of TRIM44 suppresses the invasion and migration of SKOV3 and A2780 cells in vitro
In the Transwell invasion assay, we observed that invasion was inhibited in lenti-shTRIM44-transfected cells. The migration experiment showed that knockdown of TRIM44 reduced the migration of lenti-shTRIM44 virus-transfected cells (Figure 2E,2F,2H). The migration of SKOV3 cells transfected with TRIM44 shRNA was significantly inhibited compared to that of control cells (0.115±0.002 versus 0.258±0.002, P<0.001). The migration of A2780 cells transfected with TRIM44 shRNA was also significantly inhibited (0.030±0.003 versus 0.086±0.006, P<0.001). The invasion experiment revealed that knockdown of TRIM44 reduced the invasion of lenti-shTRIM44 virus-transfected cells (Figure 2E-2G). The invasion of SKOV3 cells transfected with TRIM44 shRNA was significantly inhibited compared to that of control cells (0.100±0.004 versus 0.289±0.005, P<0.001). Invasion of A2780 cells transfected with TRIM44 shRNA was significantly inhibited (0.308±0.007 versus 0.497±0.004, P<0.001). These results indicate that lentiviral vectors containing TRIM44 shRNA effectively inhibit migration and invasion induced by TRIM44.
Knockdown of TRIM44 induces apoptosis of SKOV3 and A2780 cells in vitro
We next investigated potential cellular apoptosis events during cell death caused by TRIM44 knockdown in the SKOV3 and A2780OC cell lines. The results revealed that apoptosis levels were increased in both the SKOV3 and A2780 cell lines in response to transfection with lenti-shTRIM44 (Figure 2I,2J). Depletion of TRIM44 caused drastic morphological changes and a substantial decrease in cell number. Taken together, these results clearly indicate that TRIM44 depletion leads to apoptotic cell death in SKOV3 and A2780 cancer cells.
Knockdown of TRIM44 significantly inhibits colony-forming ability in SKOV3 and A2780 cells
The long-term effect of TRIM44 silencing on the colony-forming ability of SKOV3 and A2780 cells was assessed using colony formation assays. The size of the independent colonies was much smaller in shTRIM44-treated cells than in the nontreated and shNC-treated cells. Moreover, the number of colonies formed by SKOV3 and A2780 cells was significantly decreased following TRIM44 knockdown (P<0.05) (Figure 2K,2L). These data indicate that TRIM44 knockdown also significantly inhibits colony formation in SKOV3 and A2780OC cells.
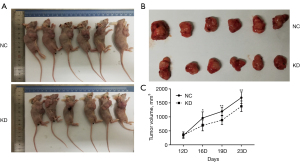
Knockdown of TRIM44 inhibits in vivo xenograft tumor growth in nude mice
To evaluate the effect ofTRIM44 on ovarian carcinogenesis in vivo, we established xenograft tumor models in nude mice using A2780 cells transfected with lenti-shTRIM44 treatment. Xenograft tumors were observed in the nude mice at the injection site, and tumors were harvested 30days after injection. The tumor volumes were measured once a week. The volume of xenograft tumors is a common criterion for judging the growth of OC cells in vivo. As shown in Figure 3, tumor volumes in the lenti-shTRIM44-transfected cell treatment group were significantly decreased compared to those in the control group (Figure 4, P<0.05). These results indicate that lentiviral vectors with the TRIM44 shRNA sequence inhibit xenograft tumor growth, indicating that downregulation of TRIM44 may be beneficial for the treatment of OC.

Global gene expression analysis following TRIM44 knockdown revealed modulation of key pathways in EOC cells
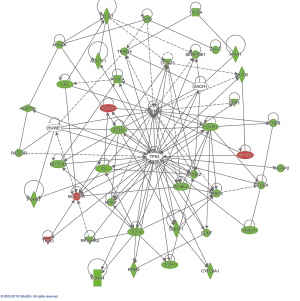
To elucidate the molecular mechanisms by which TRIM44 contributes to the carcinogenesis of OC cells, we performed gene chip analysis (gene expression profiling, using the Human Gene Expression Array PathArrayTM platform) comparing the gene expression of SKOV3 NS cells versus SKOV3 shTRIM44 cells. Differentially expressed genes with at least a 1.2-fold change were identified. Common differentially expressed genes were analyzed using ingenuity pathway analysis (IPA). We identified 1,172 genes with significant expression changes (>1.2-fold with adjusted P<0.05) (data not shown in this paper). The bioinformatics analysis showed that expression of 436 genes was significantly increased (P<0.05) while that of 736 genes was significantly decreased (P<0.05). The most markedly altered genes were AKT1S1 (AKT1 substrate 1, decreased1.96-fold), USP14 (ubiquitin specific peptidase 14, decreased1.85-fold), STK38L (serine/threonine kinase 38 like, decreased1.83-fold), GLIPR1 (GLI pathogenesis-related 1, increased1.84-fold), SEC23A (Sec23 homolog A, COPII coat complex component, decreased 1.76-fold), and SPC24 (NDC80 kinetochore complex component, decreased1.72-fold). Many of these TRIM44-regulated differentially expressed genes in EOC cells are involved in regulating cell proliferation, wound healing, the cell cycle, DNA damage, and cellular assembly and organization. In addition, functional analysis of the genes using IPA revealed that differentially expressed genes were primarily enriched in the superpathway of cholesterol biosynthesis (ACAT2, FDPS, IDI1, LBR, LSS and MVD, etc.), the superpathway of serine and glycine biosynthesis I (PHGDH, PSAT1, PSPH, and SHMT2), cyclins and cell cycle regulation (CCNA1, CCNA2, CCNB2, CCND3, CCNE2, CDK1, E2F2,E2F8, etc.), estrogen-mediated S-phase entry (CCNA1, CCNA2, CCNE2, CDK1, E2F2, etc.) the superpathway of geranylgeranyl diphosphate biosynthesis I signaling (ACAT2, FDPS, IDI1, MVD, and MVK), the NER pathway (ACAT2, FDPS, IDI1, MVD, and MVK), cell cycle control of chromosomal replication (CDC6, CDK1, CDK10, LIG1, and MCM3,etc.),and oxidative phosphorylation (COX7A1, NDUFAB1, NDUFB11, etc.). Taken together, these data indicate that TRIM44 may be a key regulator of cell proliferation and migration that controls cancer progression. Most of these pathways are implicated in tumorigenesis or chemotherapy resistance (all P<0.05; Figure 5).
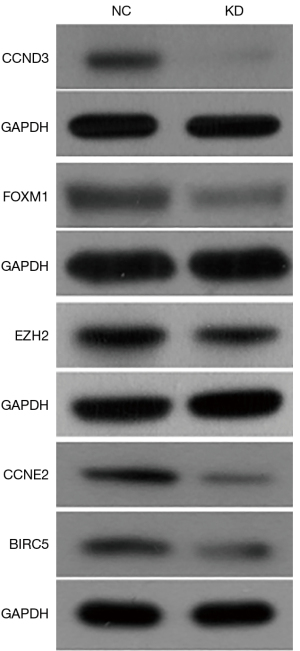
TRIM44 knockdown suppresses the FOXM1-EZH2 signaling pathway in EOC cells
The potential gene network interacting with TRIM44 was also analyzed using IPA and revealed that FOXM1-EZH2 signaling was a downstream signaling pathway regulated by TRIM44 (Figure 6). Subsequent western blot assays demonstrated that silencing TRIM44 dramatically downregulated the expression of several key proteins in the FOXM1-EZH2 signaling pathway, including FOXM1, EZH2, CCNE2, CCND3 and BIRC5 (Figure 6, P<0.05). These data further suggest that TRIM44 promotes human EOC progression via the FOXM1-EZH2 signaling pathway.
Discussion
In this study, we propose that TRIM44 acts as a metastasis-associated marker in the context of EOC cells. Knockdown of TRIM44 inhibited the invasion of EOC cells, whereas ectopic expression of TRIM44 induced this activity. Gene chip analysis further demonstrated that FOXM1-EZH2 signaling is involved in the EOC cell invasion and metastasis induced by TRIM44. Overexpression and knockdown of TRIM44 further confirmed that TRIM44 regulates invasion/metastasis by modulating the FOXM1-EZH2 signaling pathway.
The TRIM family is composed of genes that encode proteins containing TRIM. The integrated module comprises three different types of domains: a RING domain (R), a B-box domain (B), and a coiled-coil (CC) region (RBCC). The TRIM protein family is involved in a wide range of biological processes, such as cell growth, development, and cellular differentiation (14,15). Increasing evidence has shown that many TRIM family members are involved in the oncogenesis and progression of OC. For instance, TRIM52 plays an oncogenic role in the development of OC associated with the NF-kB signaling pathway (16). Furthermore, it was demonstrated that knockdown of TRIM11 not only affects the expression of apoptosis-related and invasion-related proteins but also reduces the phosphorylation levels of ERK and AKT in OC cells (17). In addition, TRIM16 inhibits migration and invasion by suppressing the Sonic hedgehog signaling pathway in OC cells (18). Given the correlation between the expression levels of these TRIMs and the tumorigenesis of OC, TRIMs represent potential novel therapeutic targets in patients with OC.
TRIM44 is one of the members of the TRIM family, which has been demonstrated to have multifaceted roles in cancer progression. Multiple reports have confirmed that TRIM44 is overexpressed in several human cancers and may promote tumor growth, proliferation and invasion (19). Kashimoto et al. showed that knockdown of TRIM44 expression inhibited the proliferation, migration, and invasion of TRIM44-overexpressing gastric cancer cells (10). Another study identified and validated that TRIM44 is independently associated with poor outcome and provides additional prognostic information in esophageal adenocarcinoma (20). Yamada et al. also found that TRIM44 promotes cell proliferation and migration and inhibits apoptosis in testicular germ cell tumors (8). Our previous study found that high TRIM44 protein expression was strongly correlated with advanced histological grade, high FIGO stage and lymph node metastasis in patients with EOC (12). Consistent with our previous data, the results of the present study indicated that TRIM44 is significantly expressed in OC tissue samples. Furthermore, to explore the role of TRIM44 in OC tumorigenesis, we knocked down TRIM44 in SKOV3 and A2780 cells. We successfully constructed an shRNA lentiviral vector that targeted TRIM44 and efficiently silenced the TRIM44 gene. In the present study, we demonstrated that depletion of TRIM44 in SKOV3 and A2780 cells suppressed invasion, migration, proliferation, and colony formation while inducing apoptosis. Metabolically, knockdown of TRIM44 significantly inhibited tumor growth in vivo. Thus, our study is consistent with the finding that TRIM44 is essential for malignant transformation of tumor cells. These findings suggest that TRIM44 may act as an oncogene in EOC.
It has been reported that TRIM44 is involved in cancer development and progression through various mechanisms, such as promoting TNFα-dependent phosphorylation of the p65 subunit of NF-κB and IκBα in breast cancer cells (6), inducing EMT via MAPK signaling in intrahepatic cholangiocarcinoma or activating the AKT/mTOR signal transduction pathway in human esophageal cancer (20). A previous study in human esophageal cancer demonstrated that TRIM44 promotes quiescent multiple myeloma cell occupancy and survival in the osteoblastic niche via HIF-1α stabilization (21). In addition, a recent study found that knockdown of TRIM44 inhibits the proliferation and invasion of papillary thyroid cancer cells by suppressing the Wnt/β-catenin signaling pathway. However, the underlying mechanisms of TRIM44 in OC cell invasion and metastasis remain to be fully elucidated. In our study, gene chip data were analyzed using IPA. Gene expression profiling using the Affymetrix Human Gene 1.0 ST platform identified 1172 transcripts that were significantly differentially expressed based on a P<0.05 threshold in SKOV3 EOC cells with TRIM44 knockdown compared to the control cells. Functional analysis of the genes using IPA revealed that TRIM44 knockdown modulates key pathways typically activated in cancer.
Moreover, the potential gene network interacting with TRIM44 was analyzed using IPA and revealed that FOXM1-EZH2signaling was a downstream pathway regulated by TRIM44. The FOXM1-EZH2 signaling pathway is a well-characterized signaling pathway related to the development of various cancer types (22-24). However, it is still unknown whether the key proteins of the FOXM1-EZH2 signaling pathway are functionally different or functionally redundant with respect to the carcinogenic mechanism of TRIM44. Subsequent western blot experiments confirmed that FOXM1-EZH2 signaling was repressed by TRIM44 knockdown, suggesting that TRIM44 positively regulates the carcinogenesis of OC cells through FOXM1-EZH2 signaling.
In conclusion, we revealed reduced expression of TRIM44 transcript expression in human ovarian tumor tissues relative to noncancerous ovarian tissues and inhibition of human EOC cell invasion, migration, and growth by TRIM44, at least in part through suppression of FOXM1-EZH2 signaling. Our findings identify a potential role for the TRIM44 gene in human EOC pathogenesis by activating the FOXM1-EZH2 signaling pathway. These conclusions need to be further validated in larger experiments.
Acknowledgments
We thank Dr. Huike Yang for participating in the evaluation procedures.
Funding: This work was supported by grants of Heilongjiang Provincial Natural Science Foundation of China (No. LH2020H124); The Liande Wu Science Foundation for Young Scholars of Harbin Medical University Cancer Hospital (No. WLD-QN1705); The Jingying Foundation of the Harbin Medical University Cancer Hospital (No. JY2015-04); The Medical Award Foundation Project of Beijing (No. YXJL-2021-0577-0421); The Science and Technology Innovation Medical Development Foundation Project of Beijing (No. KC2021-JF-0055-01) and The Outstanding Youth Fund of Harbin Medical University Cancer Hospital (JCQN2019-05).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2915/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2915/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2915/coif). All authors report that this work was supported by grants of Heilongjiang Provincial Natural Science Foundation of China (No. LH2020H124); the Liande Wu Science Foundation for Young Scholars of Harbin Medical University Cancer Hospital (No. WLD-QN1705); the Jingying Foundation of the Harbin Medical University Cancer Hospital (No. JY2015-04); The Medical Award Foundation Project of Beijing (No. YXJL-2021-0577-0421); and The Science and Technology Innovation Medical Development Foundation Project of Beijing (No. KC2021-JF-0055-01). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study and animal experiments were approved by the ethics board of Harbin Medical University Cancer Hospital (No. KY2021-48). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients. Experiments were performed under a project license (No. KY2021-48) granted by the ethics board of Harbin Medical University Cancer Hospital, in compliance with the “guidelines and suggestions laboratory animals” (Ministry of Science and Technology of the People’s Republic of China).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Wang H, Fang ZL, Zhang GH, et al. TRIM44, a crucial target of miR-410, functions as a potential oncogene in osteosarcoma. Onco Targets Ther 2018;11:3637-47. [Crossref] [PubMed]
- Luo Q, Lin H, Ye X, et al. Trim44 facilitates the migration and invasion of human lung cancer cells via the NF-κB signaling pathway. Int J Clin Oncol 2015;20:508-17. [Crossref] [PubMed]
- Kawabata H, Azuma K, Ikeda K, et al. TRIM44 Is a Poor Prognostic Factor for Breast Cancer Patients as a Modulator of NF-κB Signaling. Int J Mol Sci 2017;18:1931. [Crossref] [PubMed]
- Zhou Z, Liu Y, Ma M, et al. Knockdown of TRIM44 inhibits the proliferation and invasion in papillary thyroid cancer cells through suppressing the Wnt/β-catenin signaling pathway. Biomed Pharmacother 2017;96:98-103. [Crossref] [PubMed]
- Yamada Y, Takayama KI, Fujimura T, et al. A novel prognostic factor TRIM44 promotes cell proliferation and migration, and inhibits apoptosis in testicular germ cell tumor. Cancer Sci 2017;108:32-41. [Crossref] [PubMed]
- Tan Y, Yao H, Hu J, et al. Knockdown of TRIM44 Inhibits the Proliferation and Invasion in Prostate Cancer Cells. Oncol Res 2017;25:1253-9. [Crossref] [PubMed]
- Kashimoto K, Komatsu S, Ichikawa D, et al. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci 2012;103:2021-6. [Crossref] [PubMed]
- Ong CA, Shannon NB, Ross-Innes CS, et al. Amplification of TRIM44: pairing a prognostic target with potential therapeutic strategy. J Natl Cancer Inst 2014;106:dju050. [Crossref] [PubMed]
- Liu S, Yin H, Ji H, et al. Overexpression of TRIM44 is an independent marker for predicting poor prognosis in epithelial ovarian cancer. Exp Ther Med 2018;16:3034-40. [Crossref] [PubMed]
- Yu XZ, Yuan JL, Ye H, et al. TRIM44 facilitates ovarian cancer proliferation, migration, and invasion by inhibiting FRK. Neoplasma 2021;68:751-9. [Crossref] [PubMed]
- Hatakeyama S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem Sci 2017;42:297-311. [Crossref] [PubMed]
- Cambiaghi V, Giuliani V, Lombardi S, et al. TRIM proteins in cancer. Adv Exp Med Biol 2012;770:77-91. [Crossref] [PubMed]
- Yang W, Liu L, Li C, et al. TRIM52 plays an oncogenic role in ovarian cancer associated with NF-kB pathway. Cell Death Dis 2018;9:908. [Crossref] [PubMed]
- Chen Y, Sun J, Ma J. Proliferation and invasion of ovarian cancer cells are suppressed by knockdown of TRIM11. Oncol Lett 2017;14:2125-30. [Crossref] [PubMed]
- Tan H, Qi J, Chu G, et al. Tripartite Motif 16 Inhibits the Migration and Invasion in Ovarian Cancer Cells. Oncol Res 2017;25:551-8. [Crossref] [PubMed]
- Peng R, Zhang PF, Zhang C, et al. Elevated TRIM44 promotes intrahepatic cholangiocarcinoma progression by inducing cell EMT via MAPK signaling. Cancer Med 2018;7:796-808. [Crossref] [PubMed]
- Xiong D, Jin C, Ye X, et al. TRIM44 promotes human esophageal cancer progression via the AKT/mTOR pathway. Cancer Sci 2018;109:3080-92. [Crossref] [PubMed]
- Chen Z, Lin TC, Bi X, et al. TRIM44 promotes quiescent multiple myeloma cell occupancy and survival in the osteoblastic niche via HIF-1α stabilization. Leukemia 2019;33:469-86. [Crossref] [PubMed]
- Kim SH, Joshi K, Ezhilarasan R, et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem Cell Reports 2015;4:226-38. [Crossref] [PubMed]
- Harzer K, Yildiz Y. High β-glucosidase (GBA) activity not attributable to GBA1 and GBA2 in live normal and enzyme-deficient fibroblasts may emphasise the role of additional GBAs. Biol Chem 2015;396:1241-6. [Crossref] [PubMed]
- Perotti V, Baldassari P, Molla A, et al. An actionable axis linking NFATc2 to EZH2 controls the EMT-like program of melanoma cells. Oncogene 2019;38:4384-96. [Crossref] [PubMed]


