
LncRNA B4GALT1-AS1 promotes non-small cell lung cancer cell growth via increasing ZEB1 level by sponging miR-144-3p
Introduction
Cancer of the lungs is the most frequently occurring type of malignancy in humans and is a major reason for death due to cancer globally; about 1.35 million novel cases are diagnosed each year worldwide (1,2). Non-small cell lung cancer (NSCLC) is aggressive and exhibits a high rate of recurrence and metastasis (3). There has been no significant increase in the 5-year survival rate of patients with NSCLC, and the overall 5-year survival rate of these patients is 10% (3). As a result, it is essential to develop new therapeutic strategies that are more effective for NSCLC patients.
While long noncoding ribonucleic acids (lncRNAs; 200 bases long) are not capable of translating proteins, their dysregulation has been observed in several cancers, such as NSCLC (4-8). For example, the process of epithelial-mesenchymal transition (EMT) is enhanced by lncRNA FEZF1 antisense RNA 1 via E-cadherin suppression and Wnt pathway regulation in NSCLC (9). Furthermore, a significantly enhanced level of LINC00968 was observed in NSCLC tissues while its silencing impeded the progression of NSCLC by Wnt signaling pathway activation (10). As the antisense equivalent of B4GALT1, a significant complementarity of lncRNA B4GALT1-AS1 was observed through B4GALT1 messenger RNA (mRNA) and exhibits specific, cancer-based variations (11). The association of B4GALT1-AS1 with hnRNPA1 in attenuating glucose/lipid metabolism in the liver was recently shown (12). Nevertheless, the role and inherent mechanisms of B4GALT1-AS1 in human NSCLC development remains to be assessed. We examined the expression of B4GALT1-AS1 in NSCLC cell lines and tissues, the effect of B4GALT1-AS1 downregulation on the growth of NSCLC cell lines. Additionally, we also examined the role of lncRNA B4GALT1-AS1 in the aggressive phenotypes of NSCLC cells by using miR-144-3p binding and assessing the ZEB1 expression. Finally, we attempted to elucidate the role of the regulatory network of lncRNA B4GALT1-AS1/miR-144-3p /ZEB1 in NSCLC progression. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-296/rc).
Methods
Tissues and cell lines for NSCLC
The cell lines for NSCLC (A549, H1299, H1975, and HCC827), BEAS-2B (the human cell line from bronchial epithelium), and the 293T cell line were provided by the Nanjing KeyGen Biotech Co., Ltd., in Nanjing (China). The maintenance of cell lines for NSCLC was done in the 1640 media with 10% fetal bovine serum (FBS) from Invitrogen (Carlsbad, CA, USA), and 100 µg/mL each of streptomycin plus penicillin. The LHC-8 medium from Invitrogen was used to culture the BEAS-2B cells. Each cell line was maintained in a 37 ℃ and 5% CO2. A total of 36 NSCLC cases and equivalent adjacent tissues from lungs were sampled from the First Affiliated Hospital of Xingtai Medical College (Table 1). Before the experiments commenced, written informed consent was obtained from patients. Immediate freezing of tissues was done in liquid N2, and the tissues were kept at −80 ℃. None of the patients had undergone any treatments prior to the surgical procedure. Approval of the study was given by the Ethics Committee of the First Affiliated Hospital of Xingtai Medical College. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Characteristics | Cases |
|---|---|
| Age (years) | |
| ≥60 | 18 |
| <60 | 18 |
| Sex | |
| Male | 10 |
| Female | 26 |
| Tumor size (mm) | |
| ≤3 | 28 |
| >3 | 8 |
| Lymph node metastasis | |
| Yes | 14 |
| No | 22 |
| Tumor stage | |
| I–II | 20 |
| III–IV | 16 |
NSCLC, non-small-cell lung cancer.
Transfections of cells
To target the B4GALT1-AS1 (shB4GALT1-AS1) and NC or shCon (negative control) vector, lentivirus-mediated short hairpin RNA (shRNA) was procured from Genechem in Shanghai (China). The mimic of miR-144-3p, miR-NC, the negative control, inhibitors of miR-144-3p, and miRNA (miR-NC inhibitor) were prepared and provided by Genechem in Shanghai. In addition, lipofectamine 3000 from Invitrogen was used for cell transfections.
Assay for real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
RNA was isolated using TRIzol from Invitrogen and performed as previous (23). GAPDH was used as the reference mRNA and U6 was used as the reference miRNA genes.
Assay for cell proliferation
Cells of NSCLC were added (2×103 cells per well) into plates of 96 wells. After 12, 24, 36, or 48 full hours, the addition of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (20 µL) was done into wells and kept for 20 minutes with Dimethyl sulfoxide (DMSO) (200 µL). The estimation of optical density (OD) values was made at 490 nm on an Analyzer for Enzyme Immunoassays from Bio-Rad Laboratories (CA).
Reporter assay for luciferase
Luciferase reporter assay was performed as previous (12).
Assay for RNA immunoprecipitation (RIP)
For the RIP evaluation, the RNA-binding protein immunoprecipitation Magna RIP kit from Millipore was used. AGO2 and IgG antibodies were obtained from Abcam (Cambridge, MA, USA). The beads were washed using the wash buffer. To remove proteins, the incubation of complexes was done with sodium dodecyl sulfate (SDS) (0.1%)/proteinase K. Using the precipitated RNA, qRT-PCR assay was done to show the binding targets.
Statistical analysis
The analysis of statistical data was conducted using SPSS 20.0 (IBM Corp., New York, USA) and stated as the mean ± standard deviation (SD). Variation among the two groups was assessed through either one-way Analysis of Variance (ANOVA) or a student’s t-test (two-tailed) and the post-hoc Dunnett’s test. The Pearson correlation analysis was used to assess the miR-144-3p and B4GALT1-AS1 correlation. A P value of less than 0.05 was deemed statistically significant.
Results
Up-regulation of B4GALT1-AS1 in human NSCLC
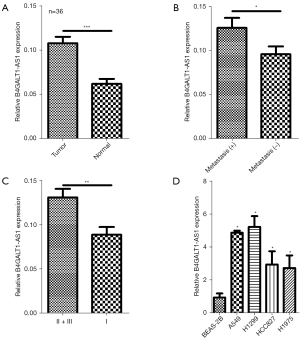
From a total of 36 cases, tissues of NSCLC and equivalent healthy tissues were sampled, and B4GALT1-AS1 expression was examined through qRT-PCR. Remarkable overexpression of B4GALT1-AS1 was observed in tissues of NSCLC than that in adjacent (normal) tissue (Figure 1A). Further enhanced B4GALT1-AS1 expression was closely associated with NSCLC metastasis and an advance stage of tumor node metastasis (Figure 1B,1C). Finally, overregulation of B4GALT1-AS1 was also observed in NSCLC cells (A549, H1299, HCC827, and H1975) in contrast to the control (BEAS-2B) cells (Figure 1D). As a result, there was an over-regulation of B4GALT1-AS1 expression in NSCLC.
Growth of NSCLC cell are promoted by B4GALT1-AS1
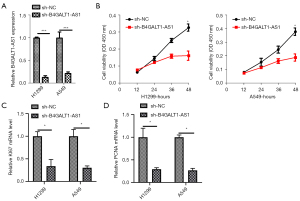
To examine the role of B4GALT1-AS1 in NSCLC progression, we used siB4GALT1-AS1 (siRNA targeting B4GALT1-AS1) and knocked-down B4GALT1-AS1 in H1299 and A549 cells (Figure 2A). Then, verification of B4GALT1-AS1 was done through qRT-PCR. The viability of H1299 and A549 cells was impeded by B4GALT1-AS1 silencing (Figure 2B). Afterwards, nocked-down B4GALT1-AS1 in H1299 and A549 cells decreased mRNA level of KI67 and PCNA mRNA level in NSCLC cells (Figure 2C,2D).
The binding of LncRNA B4GALT1-AS1 with miR-144-3p in NSCLC cell
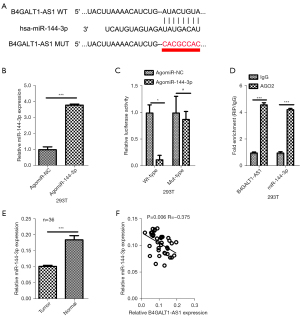
The starBase tool (http://starbase.sysu.edu.cn/) was used to screen the potential miRNAs that interacted with B4GALT1-AS1, and found that miR-144-3p as potential miRNA of B4GALT1-AS1 (Figure 3A,3B). Subsequently, co-transfection of the pmirGLO reporter vector with either B4GALT1-AS1-WT (WT B4GALT1-AS1) or B4GALT1-AS1-MUT (MUT) combined with miR-144-3p was done in 293T cells. The activity of luciferase was reduced remarkably in NSCLC cells transfected with B4GALT1-AS1-WT, while the activity of luciferase miR-144-3p did not have an inhibitory effect on the activity of luciferase in B4GALT1-AS1-MUT transfected cells (Figure 3B,3C). Next, the miR-144-3p and B4GALT1-AS1 relationship was confirmed through the RIP assay. The miR-144-3p and B4GALT1-AS1 combination complex precipitated in Ago2 (Figure 3D). The NSCLC tissues expressed miR-144-3p, which was more repressed in tissues of cancer compared to normal samples (Figure 3E). Finally, Spearman’s correlation analysis of NSCLC tissues showed an inverse relationship between B4GALT1-AS1 and miR-144-3p levels (Figure 3F). Results showed that B4GALT1-AS1 was negatively regulated by binding with miR-144-3p.
NSCLC cell aggressiveness is regulated by B4GALT1-AS1 via miR-144-3p/ZEB1 axis modulation
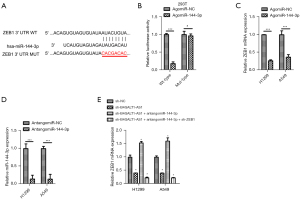
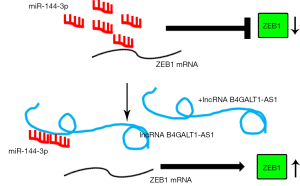
We used the bioinformatics tool, Targetscan (http://www.targetscan.org/), and detected complementary binding sites on miR-144-3p for ZEB1 (Figure 4A). After miR-144-3p transfections, the activity of luciferase in 293T cells was significantly impeded when it was transfected with ZEB1 3'-UTRWT that contained the luciferase plasmid. However, no such inhibition was observed on transfection with ZEB1 3'-UTR MUT which contained the luciferase plasmid. (Figure 4B) The qRT-PCR results further suggested that the ZEB1 level in H1299 and A549 was inhibited by miR-144-3p (Figure 4C), indicating that miR-144-3p suppressed the expression of ZEB1. Next, we decreased the expression of miR-144-3p in indicated NSCLC cell lines with miR-144-3p inhibitor (Figure 4D). The levels of ZEB1 were detected using qRT-PCR in NSCLC cells with siB4GALT1-AS1 alone, or with siB4GALT1-AS1 and the inhibitor of miR-144-3p, or by co-transfection with the si-ZEB1/miR-144-3p inhibitor/siB4GALT1-AS1 (Figure 4E). The mRNA level of NSCLC cells declined after B4GALT1-AS1 silencing. However, the effect of B4GALT1-AS1 silencing was reversed by the miR-144-3p inhibitor. Considering these results cumulatively, B4GALT1-AS1 was found to facilitate NSCLC progression through miR-144-3p/ZEB1 axis modulation (Figure 5).
Discussion
Dysregulation of several lncRNAs has been observed in the cancer of lungs, and these could also regulate the progression of NSCLC malignancy by serving as cancer oncogenes or suppressors. Several malignant tumors exhibit dysregulated lncRNAs. For example, lncRNAUCA1 regulates miR-193a-3pto enhance NSCLC progression (13). Additionally, NSCLC tissues also exhibit repressed lncRNA 00312 and correlate with poor clinical outcomes (14). Previously, lncRNA B4GALT1-AS1, the B4GALT1 antisense counterpart, was shown to have significant complementarity with B4GALT1 mRNA and displayed variations that are unique to cancer (11). B4GALT1-AS1 was found to associate with hnRNPA1 to attenuate the metabolism of glucose/lipid in the liver (12). Nonetheless, no investigation report has demonstrated the involvement of B4GALT1-AS1 in the progression of human NSCLC.
In the present study, we demonstrate found that overregulation of lncRNA B4GALT1-AS1 was present in NSCLC patients’ clinical tissues in contrast with the equivalent adjacent (normal) tissues and BEAS-2B (healthy cell line of bronchial epithelium). The silencing of B4GALT1-AS1 suppressed the invasive, proliferative, and migratory capacities of NSCLC cell lines. Moreover, B4GALT1-AS1 bound to miR-144-3p and positively regulated the expression of ZEB1 (the functional miR-144-3p target). Finally, the current work demonstrates the participation of B4GALT1-AS1 in NSCLC progression via miR-144-3p/ZEB1 regulation. Therefore, the role of TRDG1 is oncogenic in NSCLC.
LncRNAs act as quenchers of miRNA to regulate tumor progress (15-22).For example, the specific transcript, X inactive, facilitates the angiogenesis of glioma and tumorigenicity by sequestering miR-429 (23). Additionally, we also demonstrated the binding of B4GALT1-AS1 to miR-144-3p, the level of which was related inversely to the expression of B4GALT1-AS1 in NSCLC tissue. Additionally, the transfection of miR-144-3p caused a decline in the B4GALT1-AS1 level, whereas the transfection of the inhibitor of miR-144-3p enhanced its expression. The reporter assay for luciferase activity further demonstrated the binding of miR-144-3p to the 3'-UTR of B4GALT1-AS1.
Aberrant miR-144-3p expression acts either as a suppressor of tumors or as an oncogene in several cancers. For example, connexin 43 expression is regulated by miR-144-3p, causing the suppressed formation of bones in distraction osteogenesis (24). MiR-144-3p suppresses EMT in the cancer of the stomach via PBX3 down-regulation (25), acting as a suppressor of the tumor in glioblastoma by modulating the role of c-Met (26), and serving as a tumor suppressor microRNA in HCC (hepatocellular carcinoma) to regulate HCC progression (27,28). Furthermore, miR-144-3p impedes cell migration, proliferation, and invasion in the cancer of pancreas, by targeting FosB Proto-Oncogene(FOSB), the AP-1 transcription factor subunit, which can target proline-rich protein 11 to promote arrest of cell cycle and apoptosis (29,30). The oncogenic function of miR-144-3p was shown in clear cell renal cell carcinoma by ARID1A regulation (31). Repressed miR-144-3p in cells of lung cancer facilitates lung adenocarcinoma progression by enhancing EZH2 expression (32). Our study confirmed that the target of miR-144-3p is ZEB1. Our results also show the suppression of ZEB1 by miR-144-3p in NSCLC cell lines.
Overexpression of ZEB1, which is an EMT-inducing zinc finger transcription factor, is observed in various cancers and it aids in EMT and tumor invasion, initiation, growth, and metastasis (33). Recently, lncRNAs have been implicated in the miRNA/ZEB1 axis modulation in human cancers. For instance, the lncRNA ZFAS1 could counteract miR-150 and enhance the expression of ZEB1 in HCC (34). The lncRNA PTAR participates in the malignant transformation and EMT of serous cells of ovarian cancer through miR-101-3p/ZEB1 axis interaction (35). Our study shows that ZEB1 expression is enhanced by B4GALT1-AS1 in NSCLC cell lines by sequestering endogenous miR-144-3p.
Conclusions
Our study demonstrates the overexpression of lncRNA B4GALT1-AS1 in NSCLC of humans. The advancement and tumorigenesis of NSCLC cell lines were promoted by B4GALT1-AS1 because of miR-144-3p sponging and because miR-144-3p positively regulated the expression of ZEB1.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-296/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-296/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-296/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Before the experiments commenced, written informed consent was obtained from patients. Approval of the study was given by the Ethics Committee of the First Affiliated Hospital of Xingtai Medical College. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Primers 2015;1:15009. [Crossref] [PubMed]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [Crossref] [PubMed]
- Chheang S, Brown K. Lung cancer staging: clinical and radiologic perspectives. Semin Intervent Radiol 2013;30:99-113. [Crossref] [PubMed]
- Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol 2011;22:366-76. [Crossref] [PubMed]
- Delás MJ, Hannon GJ. lncRNAs in development and disease: from functions to mechanisms. Open Biol 2017;7:170121. [Crossref] [PubMed]
- Pan J, Tang Y, Liu S, et al. LIMD1-AS1 suppressed non-small cell lung cancer progression through stabilizing LIMD1 mRNA via hnRNP U. Cancer Med 2020;9:3829-39. [Crossref] [PubMed]
- Ying J, Yang J, Liu Y. LncARSR promotes non-small-cell lung cancer progression via regulating PTEN/Akt. Am J Transl Res 2020;12:857-66. [PubMed]
- Wei S, Liu J, Li X, et al. LncRNA MIR17HG inhibits non-small cell lung cancer by upregulating miR-142-3p to downregulate Bach-1. BMC Pulm Med 2020;20:78. [Crossref] [PubMed]
- He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC). Biomed Pharmacother 2017;95:331-8. [Crossref] [PubMed]
- Wang Y, Zhou J, Xu YJ, et al. Long non-coding RNA LINC00968 acts as oncogene in NSCLC by activating the Wnt signaling pathway. J Cell Physiol 2018;233:3397-406. [Crossref] [PubMed]
- Al-Obaide MA, Alobydi H, Abdelsalam AG, et al. Multifaceted roles of 5'-regulatory region of the cancer associated gene B4GALT1 and its comparison with the gene family. Int J Oncol 2015;47:1393-404. [Crossref] [PubMed]
- Li Z, Wang Y, Hu R, et al. LncRNA B4GALT1-AS1 recruits HuR to promote osteosarcoma cells stemness and migration via enhancing YAP transcriptional activity. Cell Prolif 2018;51:e12504. [Crossref] [PubMed]
- Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett 2016;371:99-106. [Crossref] [PubMed]
- Zhu Q, Lv T, Wu Y, et al. Long non-coding RNA 00312 regulated by HOXA5 inhibits tumour proliferation and promotes apoptosis in Non-small cell lung cancer. J Cell Mol Med 2017;21:2184-98. [Crossref] [PubMed]
- Zou Y, Shen C, Shen T, et al. LncRNA THRIL is involved in the proliferation, migration, and invasion of rheumatoid fibroblast-like synoviocytes. Ann Transl Med 2021;9:1368. [Crossref] [PubMed]
- Zhang Z, Nong L, Chen ML, et al. Long Noncoding RNA SNHG10 Sponges miR-543 to Upregulate Tumor Suppressive SIRT1 in Nonsmall Cell Lung Cancer. Cancer Biother Radiopharm 2020;35:771-5. [Crossref] [PubMed]
- Gao L, Chen X, Wang Y, et al. Up-Regulation of FSTL3, Regulated by lncRNA DSCAM-AS1/miR-122-5p Axis, Promotes Proliferation and Migration of Non-Small Cell Lung Cancer Cells. Onco Targets Ther 2020;13:2725-38. [Crossref] [PubMed]
- Wei CM, Zhao XF, Qiu HB, et al. The long non-coding RNA PVT1/miR-145-5p/ITGB8 axis regulates cell proliferation, apoptosis, migration and invasion in non-small cell lung cancer cells. Neoplasma 2020;67:802-12. [Crossref] [PubMed]
- Li K, Jiang Y, Xiang X, et al. Long non-coding RNA SNHG6 promotes the growth and invasion of non-small cell lung cancer by downregulating miR-101-3p. Thorac Cancer 2020;11:1180-90. [Crossref] [PubMed]
- Li S, Mei Z, Hu HB, et al. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol 2018;233:6679-88. [Crossref] [PubMed]
- Wu XF, Lu JT, Chen W, et al. Mechanism of LncRNA FOXC2-AC1 promoting lung cancer metastasis by regulating miR-107. Eur Rev Med Pharmacol Sci 2019;23:690-8. [PubMed]
- Yang B, Zhang L, Cao Y, et al. Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer 2017;16:154. [Crossref] [PubMed]
- Cheng Z, Li Z, Ma K, et al. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer 2017;8:4106-16. [Crossref] [PubMed]
- Sun YX, Zhang JF, Xu J, et al. MicroRNA-144-3p inhibits bone formation in distraction osteogenesis through targeting Connexin 43. Oncotarget 2017;8:89913-22. [Crossref] [PubMed]
- Li B, Zhang S, Shen H, et al. MicroRNA-144-3p suppresses gastric cancer progression by inhibiting epithelial-to-mesenchymal transition through targeting PBX3. Biochem Biophys Res Commun 2017;484:241-7. [Crossref] [PubMed]
- Lan F, Yu H, Hu M, et al. miR-144-3p exerts anti-tumor effects in glioblastoma by targeting c-Met. J Neurochem 2015;135:274-86. [Crossref] [PubMed]
- Wu M, Huang C, Huang X, et al. MicroRNA-144-3p suppresses tumor growth and angiogenesis by targeting SGK3 in hepatocellular carcinoma. Oncol Rep 2017;38:2173-81. [Crossref] [PubMed]
- Liang HW, Ye ZH, Yin SY, et al. A comprehensive insight into the clinicopathologic significance of miR-144-3p in hepatocellular carcinoma. Onco Targets Ther 2017;10:3405-19. [Crossref] [PubMed]
- Liu S, Luan J, Ding Y. miR-144-3p Targets FosB Proto-oncogene, AP-1 Transcription Factor Subunit (FOSB) to Suppress Proliferation, Migration, and Invasion of PANC-1 Pancreatic Cancer Cells. Oncol Res 2018;26:683-90. [Crossref] [PubMed]
- Li J, Sun P, Yue Z, et al. miR-144-3p Induces Cell Cycle Arrest and Apoptosis in Pancreatic Cancer Cells by Targeting Proline-Rich Protein 11 Expression via the Mitogen-Activated Protein Kinase Signaling Pathway. DNA Cell Biol 2017;36:619-26. [Crossref] [PubMed]
- Xiao W, Lou N, Ruan H, et al. Mir-144-3p Promotes Cell Proliferation, Metastasis, Sunitinib Resistance in Clear Cell Renal Cell Carcinoma by Downregulating ARID1A. Cell Physiol Biochem 2017;43:2420-33. [Crossref] [PubMed]
- Liu C, Yang Z, Deng Z, et al. Downregulated miR-144-3p contributes to progression of lung adenocarcinoma through elevating the expression of EZH2. Cancer Med 2018;7:5554-66. [Crossref] [PubMed]
- Caramel J, Ligier M, Puisieux A. Pleiotropic Roles for ZEB1 in Cancer. Cancer Res 2018;78:30-5. [Crossref] [PubMed]
- Li T, Xie J, Shen C, et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res 2015;75:3181-91. [Crossref] [PubMed]
- Liang H, Yu T, Han Y, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer 2018;17:119. [Crossref] [PubMed]
(English Language Editor: C. Mullens)


