Bioinformatics analysis of potential glioblastoma circular RNA sponge network
Introduction
Glioma is most common intracranial tumor in adults. And among different types of glioma, glioblastoma multiforme (GBM) is the most malignant with rapid progress and poor prognosis. Through multi omics and epigenetics studies of glioma, researchers found enormous gene differences and key genetic changes between glioma and normal brain: such as TP53 and IDH mutations. Some of these differentially expressed genes are oncogenes, which promote the occurrence, development, metastasis, and invasion of tumors; while others tumor suppressor genes, which inhibit the proliferation and promote apoptosis. However, the biological significance and regulatory mechanism of these genes are not totally clear.
Circular RNA (circRNA) attribute to noncoding RNA. Due to back split mechanism, the sheared linear RNA will shape closed-loop structure which can contain several parts of sequences. Some circRNA sequences may contain exons or introns. The closed-loop structure maintains its stability not degraded by RNA exonuclease. Rybak-Wolf et al. (1) discovered that mammalian brain tissue is rich of circRNAs. However, circRNAs’ functions in human brain still remain unclear. Some scientists have discovered that circRNA can regulate genes expression. The mechanisms include “molecular-sponge” (2-4), direct translation (5) and absorbing RNA binding proteins (RBD) (6,7).
In recent years, Hansen and his colleagues proposed a “molecular sponge” regulation mechanism between RNAs (4,8). According to the base complementary pairing principle, the certain nucleotides arrangement of circRNA or microRNA can combine with other RNAs (messenger RNA, message RNA, mRNA). The full length of circRNA or microRNA sequences are often much longer than the length of binding sites. CircRNA and microRNA molecule always have multiple binding sites. Therefore, they can absorb many other RNAs (such as microRNA or mRNA), which is like the absorption of water by sponge. In essence, it is also a competitive endogenous RNA (ceRNA) mechanism (2). This effect may come into extensive and powerful regulatory of downstream pathways, so that affect a variety of biological processes such as tumor proliferation or invasion.
This study analyzed the sequencing and array data of multiple GBM circRNA datasets in Gene Expression Omnibus (GEO) database and other data in several large international and Chinese databases. We selected the common differentially expressed circRNA and microRNA to construct the circRNA-microRNA- gene network, in order to explore new targets for primary GBM comprehensively and accurately.
Methods
Overall design
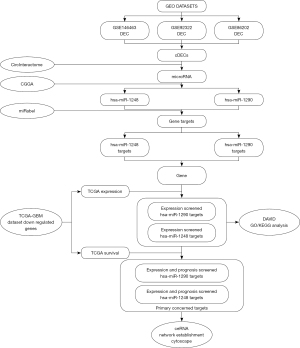
This study firstly searched and screened differentially expressed circRNAs (DECs) from GEO datasets. Then, a circRNA-microRNA interaction database was used to predict potential microRNA targets. These potential microRNAs would be screened by microRNA expression database to ensure the expression trend accords with “ceRNA theory”. After that the screened microRNAs’ probable gene targets would be calculated by a microRNA-gene interaction database. At the same time, we extracted the differentially expressed genes (DEGs) of glioblastoma compared with normal brain tissue from The Cancer Genome Atlas (TCGA) database. Next, we figured out the specific TCGA DEGs in microRNA targets whose expression accord with “ceRNA theory” equally. Last, both potential microRNAs and their DEG targets were re-screened by survival analysis to establish a possible “Molecular Sponge Network”. The function enrichment analysis of DEGs with prognosis significant was also induced by Database for Annotation, Visualization and Integrated Discovery (DAVID) and Kyoto Encyclopedia of Genes and Genomes (KEGG). The analysis process is shown in Figure 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differentially expressed circRNAs (DEC) data selection
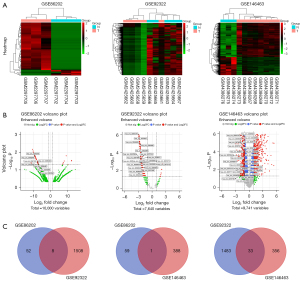
We screened the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/gds/) database by setting the search strategy as “circRNA” and “glioblastoma”. The inclusion criteria were as follows: (I) datasets contain primary glioblastoma data; (II) circRNA data is essential; (III) both GPL probe sequence files and raw data of sample sequencing are available. The exclusion criteria were: (I) data contain recurrent glioblastoma samples; (II) data of children samples; (III) no circRNA data or data not available. After dataset inclusion, differentially expressed circRNAs of datasets were extracted. Considering the different identification methods of sequencing platform, converting the circRNA identities from different GEO datasets is imperative for further analysis. CircAtlas 2.0 website (http://circatlas.biols.ac.cn/) integrates multiple databases and converts among the circRNA chromosome location, circAltas, circBase, circRNAdb, deepbase2, circpedia2 and other databases IDs. We choose circBase ID format to annotate GSE86202 and GSE92322 DECs. Arrystar platform probe annotation was used for GSE146463 DEC converting.
Prediction and screening of circRNA targets
- CircInteractome calculate potential microRNAs targets of circRNA. In order to further explore the microRNAs regulated by circRNAs, we ran the CircInteractome database and use DECs’ circBase ID (https://circinteractome.irp.nia.nih.gov/) (9) to predict circRNA microRNA interaction.
- Screening. We used Chinese Glioma Gene Atlas mapping program (CGGA, http://www.cgga.org.cn/analyse) to screen the microRNAs. CGGA is a glioma and GBM mRNA and microRNA expression, prognosis and methylation database developed by Jiang Tao team of Beijing Tiantan Hospital (10). When screening, the first step is to analyze the microRNA distribution in different grades of glioma. According to “ceRNA theory”, microRNA expression trend must be opposite to circRNA, which means down-regulated circRNA must correspond to up-regulated microRNA, vice versa. MicroRNAs expression data were from CGGA _array_198 chip, which contains 198 different glioma patients’ microRNA data. MicroRNAs expression difference must be significant in both overall glioma and glioblastoma (WHO grade IV). After expression screening, the microRNA prognosis analysis was also taken into consideration.
After screening, the primary circRNA-microRNA interactions were determined. The sequence length, binding site of DECs and microRNA targets were extracted from circBase and CircInteractome databases to visualize the binding sites between circRNAs and microRNAs.
Prediction and screening of microRNA targets
MiRabel (http://bioinfo.univ-rouen.fr/mirabel/) (11) is a powerful online interaction tool integrating gene targets data from MiRanda (http://www.microrna.org/microrna/home.do), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), SVmicro (http://compgenomics.utsa.edu/svmicro.html), TargetScan (http://www.targetscan.org/vert_80/) prediction tools and miRwalk database (http://mirwalk.umm.uni-heidelberg.de/) and microRNA data from miRbase (https://www.mirbase.org/) to automatically predict possible targets. The testing threshold was set as P<0.05. The result would be screened by PITA, miRanda, SVMicro and Targetscan scores. Only all predicting scores of these four databases were available, the predicted targets could be selected.
Screening of genes by TCGA
The microRNA targets need not only screened by algorithm but also tested by real-world data. The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) is the largest and authoritative database of tumor research. Containing enormous clinical and sequencing data of various tumor samples, TCGA could filter the significant difference of gene targets commendably. Primary glioblastoma samples sequencing data and corresponding clinical information of TCGA-GBM project was downloaded, extracted, and analyzed. Differentially expressed genes (DEGs)heat-map and volcano map were also calculated. Then genes predicted by miRabel were rescreened by TCGA-GBM DEGs and survival data.
At last, in order to accord with “ceRNA” theory, screened genes could be divided into two circumstances: (I) if microRNAs were up-regulated and with poor prognosis, the gene targets of microRNAs should be down-regulated with good prognosis; (II) if microRNAs were up-regulated with good prognosis, the gene targets of microRNAs should be down-regulated with poor prognosis.
Functional enrichment analysis
Database for annotation, visualization and integrated discovery (DAVID, https://david.ncifcrf.gov/) V6.8 database is used for gene ontology (GO) analysis and Kyoto Encyclopedia of genes and genomes (KEGG) analysis. First, the selected gene list is identified as “OFFICIAL_GENE_SYMBOL”; “Homo Sapiens” and “GENE LIST” for further analysis. Then, the “Analyze above gene list with one of DAVID tools” and “Functional annotation tool” were applied to get the annotation summary results of “GO term-BP”, “GO term-CC”, “GO term-MF” and “KEGG-pathways”.
Construction of molecular sponge network
The DECs, screened microRNAs, and screened genes’ potential molecular sponge interactions were visualized by Cytoscape 3.9.0 software. Gene targets with prognosis significance were labeled specially.
Statistical analysis
Test level of DECs and DEGs were set as |logFoldChange (logFC)|>1 and P<0.05 by “edgeR” with R 4.03 software (12). Log-rank test, univariate Cox proportional hazards regression, P value, 95% confidence interval (CI) and hazard ratio (HR) were used for survival assessment. HR >1 means high expression of gene associated with worse prognosis and 0< HR <1 referred to better prognosis. P<0.05 was considered as statistically significant.
P<0.05 is also set as the significant threshold for GO and KEGG enrichment analysis.
Visualization
The Kaplan Meier curve was induced by “survival” and “surviviner” packages. The “ggplot2” package output graphical results. DEC for each GEO dataset was visualized as heat-map and volcano-plot. DECs were also induced intersection to discover common DECs (cDECs) by Venn plot online server (http://bioinformatics.psb.ugent.be/webtools/Venn/). The visualization of binding can be fulfilled by bioinformatics online mapping tool (http://www.bioinformatics.com.cn).
Results
Acquisition of differentially expressed circRNAs
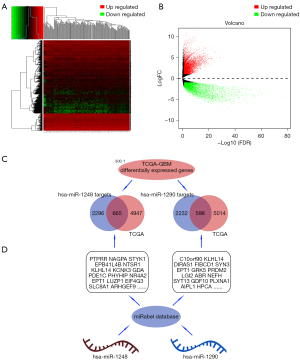
The keywords “GBM” and “circRNA” were searched in the GEO database. After screening the results, we finally got four datasets: GSE146463 by Bronisz et al. (13), GSE92322 by Zhu et al. (14), GSE86202 by Yuan et al. (15), and GSE153692. Because of being unable to be recognized by circAtlas conversion system, GSE153692 dataset is excluded from analysis. All differentially expressed circRNAs were extracted by “edgeR” package. All FDR values were 0.807043219 in GSE86202. And 752 FDR >0.05 were detected in GSE92322. And 1,672 DEC FDR >0.05 in GSE146463 (available online: https://cdn.amegroups.cn/static/public/tcr-21-2597-01.zip). If we test with the FDR<0.05 level, large number of potential circRNAs might be ignored. So we set |logFC|>1, P<0.05 as filter condition. Excluding circRNAs not being annotated with circBase ID, 737 up-regulated and 389 down-regulated DECs of GSE146463, 0 up-regulated and 1,516 down-regulated DECs of GSE92322 and 7 up-regulated and 60 down-regulated DECs of GSE86202 were obtained. No up-regulated common DEC (cDEC) was discovered among the three datasets. There were 42 down-regulated cDECs. GEO datasets profile, and cDECs’ details were shown in Tables 1,2 and Figure 2.
Table 1
| Dataset ID | CircRNAs (n) | Sample (n) | DECs (n) | Annotated DECs (n) | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | N | Total | Up | Down | Total | Up | Down | |||||
| GSE86202 | 37,084 | 3 | 3 | 6 | 9 | 149 | 158 | 7 | 60 | 67 | ||
| GSE92322 | 59,386 | 5 | 5 | 10 | 11 | 1,995 | 2,006 | 0 | 1,516 | 1,516 | ||
| GSE146463 | 12,660 | 8 | 3 | 11 | 822 | 539 | 1,361 | 737 | 389 | 1,126 | ||
DEC, differentially expressed circRNAs; GEO, Gene Expression Omnibus; GSE, Gene Expression Omnibus Series; ID, identity.
Table 2
| Intersection | DEC number | DEC ID |
|---|---|---|
| GSE146463-down & GSE86202-down | 1 | hsa_circ_0002096 |
| GSE146463-down & GSE92322-down | 33 | hsa_circ_0005654 |
| hsa_circ_0070113 | ||
| hsa_circ_0035957 | ||
| hsa_circ_0006956 | ||
| hsa_circ_0079557 | ||
| hsa_circ_0034182 | ||
| hsa_circ_0004619 | ||
| hsa_circ_0000219 | ||
| hsa_circ_0058495 | ||
| hsa_circ_0007848 | ||
| hsa_circ_0003390 | ||
| hsa_circ_0005566 | ||
| hsa_circ_0001073 | ||
| hsa_circ_0007771 | ||
| hsa_circ_0006499 | ||
| hsa_circ_0047880 | ||
| hsa_circ_0030788 | ||
| hsa_circ_0006931 | ||
| hsa_circ_0009000 | ||
| hsa_circ_0006580 | ||
| hsa_circ_0008021 | ||
| hsa_circ_0009064 | ||
| hsa_circ_0019799 | ||
| hsa_circ_0001792 | ||
| hsa_circ_0000932 | ||
| hsa_circ_0051348 | ||
| hsa_circ_0075648 | ||
| hsa_circ_0037516 | ||
| hsa_circ_0030787 | ||
| hsa_circ_0023858 | ||
| hsa_circ_0005535 | ||
| hsa_circ_0004538 | ||
| hsa_circ_0008390 | ||
| GSE86202-down & GSE92322-down | 8 | hsa_circ_0002590 |
| hsa_circ_0008456 | ||
| hsa_circ_0070700 | ||
| hsa_circ_0032249 | ||
| hsa_circ_0069249 | ||
| hsa_circ_0047391 | ||
| hsa_circ_0069492 | ||
| hsa_circ_0024366 |
DEC, differentially expressed circRNAs; GSE, Gene Expression Omnibus Series; &, find intersection; ID, identity.
CircInteractome predicts circRNA microRNA targets
Calculated by the CircInteractome database, 761 microRNAs were predicted. Details were listed in Table 3.
Table 3
| circ ID | MicroRNA number (n) |
|---|---|
| hsa_circ_0002096 | 19 |
| hsa_circ_0005654 | 35 |
| hsa_circ_0070113 | 18 |
| hsa_circ_0035957 | 24 |
| hsa_circ_0006956 | 15 |
| hsa_circ_0079557 | 19 |
| hsa_circ_0034182 | 7 |
| hsa_circ_0004619 | 14 |
| hsa_circ_0000219 | 39 |
| hsa_circ_0058495 | 34 |
| hsa_circ_0007848 | 22 |
| hsa_circ_0003390 | 26 |
| hsa_circ_0005566 | 11 |
| hsa_circ_0001073 | 15 |
| hsa_circ_0002590 | 17 |
| hsa_circ_0008456 | 3 |
| hsa_circ_0070700 | 16 |
| hsa_circ_0032249 | 16 |
| hsa_circ_0007771 | 25 |
| hsa_circ_0006499 | 24 |
| hsa_circ_0047880 | 6 |
| hsa_circ_0030788 | 16 |
| hsa_circ_0006931 | 16 |
| hsa_circ_0009000 | 10 |
| hsa_circ_0006580 | 26 |
| hsa_circ_0008021 | 7 |
| hsa_circ_0009064 | 12 |
| hsa_circ_0019799 | 6 |
| hsa_circ_0001792 | 11 |
| hsa_circ_0000932 | 6 |
| hsa_circ_0051348 | 14 |
| hsa_circ_0075648 | 47 |
| hsa_circ_0037516 | 13 |
| hsa_circ_0030787 | 13 |
| hsa_circ_0023858 | 43 |
| hsa_circ_0005535 | 19 |
| hsa_circ_0004538 | 7 |
| hsa_circ_0008390 | 15 |
| hsa_circ_0069249 | 23 |
| hsa_circ_0047391 | 21 |
| hsa_circ_0069492 | 26 |
| hsa_circ_0024366 | 5 |
| total | 761 |
cDECs, common differentially expressed circRNAs; ID, identity.
Screening of microRNAs in CGGA database
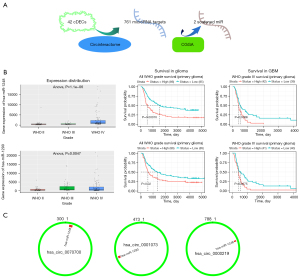
Integrated with microRNA expression, distribution, prognosis analysis and glioma WHO grades, hsa-miR-1248 and hsa-miR-1290 were screened of up-regulated and related to whole glioma (P=0.00016 for hsa-miR-1248 and P=0.02 for hsa-miR-1290) and GBM patients (P=0.0066 for hsa-miR-1248 and P=0.0075 for hsa-miR-1290) poor prognosis. Nevertheless, hsa-miR-330-5p and hsa-miR-874 associated with a worse prognosis in whole glioma (P<0.05) but better prognosis in glioblastoma (P<0.05) seemed paradox. So they were excluded. See Table 4 and Figure 3.
Table 4
| circ or miR ID | Start | End | CGGA expression | CGGA survival |
|---|---|---|---|---|
| hsa_circ_0070700 | 1 | 300 | – | – |
| hsa-miR-1248 | 199 | 205 | Up | Highly expressed with worse prognosis |
| hsa_circ_0000219 | 1 | 788 | – | – |
| hsa-miR-1248 | 151 | 157 | Up | Highly expressed with worse prognosis |
| hsa_circ_0001073 | 1 | 473 | – | – |
| hsa-miR-1290 | 326 | 332 | Up | Highly expressed with worse prognosis |
circ, circular RNA; miR, microRNA; CGGA, Chinese Glioma Genome Atlas; ID, identity.
Prediction of microRNA downstream targets
We search hsa-miR-1248, and hsa-miR-1290 potential targets in miRabel database. 2,690 gene targets of hsa-miR-1248, and 2830 gene targets of hsa-miR-1290 were predicted (P<0.05). However, whether these genes targets being significant in GBM and accords with “ceRNA” trends was unclear and worth further exploration. Only down-regulated gene targets in GBM tissues would be screened.
Gene targets screening in TCGA-GBM database
After entering TCGA database, we select the “Case” tab in “Exploration”, and “Brain” as the “Primary site”, “TCGA” in “Program”, “GBM” in the “Project”, “Gliomas” in the “Disease type”, and “primary tumor” in sample type. Through “repository” and “transcript profiling” option, the TCGA-GBM data were download. Strawberry Perl (64-bit) 5.30.0.1 software and “edgeR” package were induced to get DEGs and establish expression matrix.
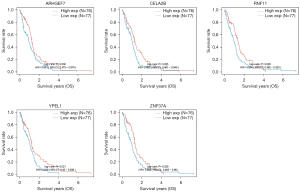
The targets of hsa-miR-1248 andhsa-miR-1290 were intersected with significantly down-regulated genes in TCGA-GBM. 643 hsa-miR-1248 targets and 570hsa-miR-1290 targets were screened with expression level. Finally, 5 genes were significantly associated with better prognosis: ARHGEF7 (log-rank P=0.036, HR =0.681, 95% CI: 0.475−0.976); CELA2b (log-rank P=0.025, HR =0.662, 95% CI: 0.461−0.949); RNF11 (log-rank P=0.026, HR =0.664, 95% CI: 0.462−0.953); YPEL1 (log-rank P=0.021, HR =0.649, 95% CI: 0.45−0.936) and ZNF37a (log-rank P=0.029, HR =0.668, 95% CI: 0.465−0.96). Details were listed in Table 5 and Figures 4,5.
Table 5
| miR ID | Targets | Screened by expression level | Screened by survival analysis | |
|---|---|---|---|---|
| Good prognosis | Bad prognosis | |||
| hsa-miR-1248 | 2,690 | 643 | 5 | 29 |
| hsa-miR-1290 | 2,830 | 570 | 3 | 21 |
TCGA, The Cancer Genome Atlas; GBM, glioblastoma multiforme; good prognosis, highly expressed targets along with good prognosis; bad prognosis, highly expressed targets along with bad prognosis; ID, identity; miR, microRNA.
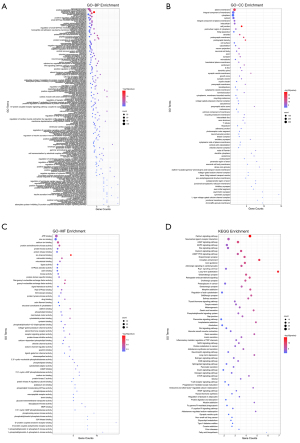
Function enrichment
All expression screened targets were analyzed by DAVID from gene ontology (GO) and KEGG pathways. GO analysis included three aspects: biological process (BP), cellular components (CC), molecular function (MF). Both GO and KEGG pathway analysis indicated that hsa-miR-1248 targets were enriched in 55 pathways, 31 pathways with significant (P<0.05), and hsa-miR-1290 targets enriched in 68 pathways, 49 pathways statistically significant (P<0.05). Sort by counts, genes were most enriched in signal transduction (counts =100, P<0.01), cell plasma membrane (counts =328, P<0.01) and ATP binding (counts =112, P<0.01) of GO and calcium signaling pathway of KEGG (counts =41, P<0.01). The prognosis associated gene targets were also checked in theses GO and KEGG pathways. For instance, ARGHEF7 was mainly enriched in signal transduction and positive regulation of apoptotic process and performed function of guanyl-nucleotide exchange and protein kinase binding. ARGHEF were also involved in regulation of actin cytoskeleton pathway. RNF11 was associated with zinc ion binding function. CELA2B was involved in pancreatic secretion and protein digestion-absorption. Details were shown in Figure 6 and available online: https://cdn.amegroups.cn/static/public/tcr-21-2597-01.zip.
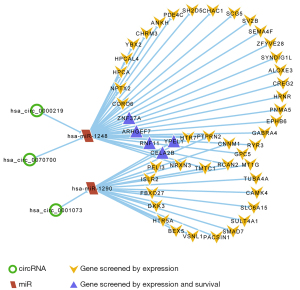
Construction of molecular sponge network of circRNA-microRNA-genes in GBM
After the extraction and screening of above databases, the eligible circRNA-microRNA-genes sponge network was visualized by Cytoscape (version 3.9.0). Hsa_circ_0000219, hsa_circ_0070700, hsa_circ_0001073, hsa-miR-1248, hsa-miR-1290 and potential downstream gene targets were particularly remarked with different shapes and colors. ARHGEF7, CELA2b, RNF11, YPEL1 and ZNF37a associated with GBM better prognosis were highlighted (Figure 7).
Discussion
In recent years, microRNA’s molecular sponge role to mRNA has been verified in glioma. Nevertheless, there is still lack of research on “circRNA-microRNA-gene” sponge network in glioma and GBM. TCGA includes enormous gene and microRNA but no circRNA expression and survival data. GEO only contains circRNA expression data. As we all know, GEO data comes from different platforms and datasets. During the ID conversion, some circRNAs had already lost because they could not be annotated as circBase IDs. We realized P value was less stringent than the FDR test. But if we insist FDR <0.05 filter standard, we would lose the whole GSE86202 dataset. So we used other conditions to filter. LogFC represents the fold change of differential gene expression, which can directly reflect the expression of circRNA. As an additional condition, more significant differential genes can be retained after logFC screening. And the shortcomings of not being tested by FDR values will partially be improved.
Moreover, single dataset analysis is difficult to ensure the data quality and the results reliability. Therefore, the joint analysis of multiple datasets could highly elevate targets screening efficiency. For thoroughly analyzing, we have included all current circRNA datasets of primary GBM. The circRNA annotation appeared diverse because of the difference between sequencing platforms and companies. For most circRNA-microRNA interaction databases using circBase ID, we also adopt circBase ID for circRNA annotation to conduct further research. However, some circRNAs cannot be identified because they do not have circBase ID. The loss of circRNAs and survival data may bring biases to results. And current reality of insufficient glioblastoma datasets makes it difficult to select more samples. The reason for selecting CGGA database to screen microRNAs is also simple: (I) TCGA microRNA only contains low grade glioma (LGG) data but lack of glioblastoma microRNA data; (II) CGGA is the first database of Chinese population. So the screening of CGGA to microRNA targets may come to conclusions fit for Chinese domestic population. For microRNA targets, we compare current large microRNA gene interaction databases including miRabel, miRanda, miRdb and RNA22 and finally choose miRabel database. MiRabel contains emerging data updated in 2020 and adpot multiple algorithms of PITA, miRanda, SVmicr0 and Targetscan. So the results can be guaranteed comprehensively and reliably (11).
Most of all, the meaning of the sponge network needs to be comprehended. In current published literature, the research rarely reported these circRNAs, microRNAs or genes on glioma or glioblastoma.
FAM188a is the host gene of hsa_circ_0000219 and also called MINDY3 (MINDY Lysine 48 Deubiquitinase 3), CARP or C10orf97. MINDY3 can activate Caspase-apoptosis and has been proven suppressor of the gastric cancer and non-small-cell lung cancer (16-18). ANK2 is the host gene of hsa_circ_0070700 whose full name is Ankyrin2. ANK2 mainly links to the integral membrane proteins and is essential for maintaining cardio-myocytes Na/Ca exchanger 1 stability (19-21). ACVR2A is the host gene of hsa_circ_0001073 associate with preeclampsia (22,23). The predicted microRNAs’ functions are highly involved in multiple tumor regulation. In particular, hsa-miR-1248 and hsa-miR-1290 show obvious oncology-genesis characteristics (24-26). However, their potential regulate function in GBM needs further research.
ARHGEF7 (Rho guanine nucleotide exchange factor 7, AK-interacting exchange factor beta) is a G protein that activates the Ras like family of Rho proteins. It plays important roles in tumor migration and metastasis (27,28). CELA2b belongs to human elastase gene of pancreatic secretion and protein digestion. However, no current reports show CELA2b can degrade desmin, vimentin or actin of tumor tissue. YPEL1 is conserved and expressed in the first half of early embryos and participates with regulating cell morphology, behavior, and development of craniofacial complex (29). ZNF37a (zinc finger protein 37a) is mainly involved in herpesvirus infection and transcriptional regulation. At present, the specific roles of these genes in GBM are not clear.
GO and KEGG is essentially a hyper-geometric distribution test for screened targets. As the effect of prior knowledge limitations cannot be ignored, the GO and KEGG results may exist bias. So we cannot deem that the gene targets and their molecular sponge axis are meaningless in glioma. More experiments are urgently needed.
Unlike those targets previously described, RNF11 (ring finger protein 11), a ubiquitin E3 ligases, is with ubiquitin protein transferase activity and forms ubiquitin editing protein complex with TNFAIP3 (A20), ITCH and TAX1BP1 to inhibit NF–κB signaling pathway over-activation and prevent tissue damage caused by inflammatory response (30,31). It has been confirmed that NF-κB signaling pathway including the classical IKK α, IKK β pathway (32) and IKBKE, TBK1/cRel pathway (33) are activated in GBM (34). NF-κB signaling pathway can promote epithelial mesenchymal transformation (EMT) (35), angiogenesis (36) and resistance to radiotherapy (37,38). The ubiquitination process mediated by RNF11 can negatively regulate and promote the termination of NF-κB signal pathway (39), which may emerge as potential anti-tumor target. RNF11 shows antitumor properties in many tumors, such as promoting ubiquitination of TGF-β and EGFR to inhibit hepatoma proliferation and migration (40). Overexpression of RNF11 also inhibits tumor proliferation in early breast cancer (41). Therefore, RNF11 regulation network is worth concerning in future research.
Conclusions
Through a series of bioinformatics analysis, we finally get the GBM circRNA sponge network. Hsa_circ_0000219, hsa_circ_070700 and hsa_circ_0001073 targeting miR-1248 and miR-1290 were of value to research, especially the function of RNF11 and its regulation network. These molecular sponge networks may bring important significance of fighting against GBM. Nevertheless, the conclusions of this research are merely database predictions, which need to be confirmed by further experiments in vivo and in vitro.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2597/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol 2018;1087:67-79. [Crossref] [PubMed]
- Lin X, Chen Y. Identification of Potentially Functional CircRNA-miRNA-mRNA Regulatory Network in Hepatocellular Carcinoma by Integrated Microarray Analysis. Med Sci Monit Basic Res 2018;24:70-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Liu XQ, Gao YB, Zhao LZ, et al. Biogenesis, research methods, and functions of circular RNAs. Yi Chuan 2019;41:469-85. [PubMed]
- Harper KL, Mcdonnell E, Whitehouse A. CircRNAs: From anonymity to novel regulators of gene expression in cancer Int J Oncol 2019;55:1183-93. (Review). [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 2016;13:34-42. [Crossref] [PubMed]
- Zhao Z, Zhang KN, Wang Q, et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genomics Proteomics Bioinformatics 2021;19:1-12. [Crossref] [PubMed]
- Quillet A, Saad C, Ferry G, et al. Improving Bioinformatics Prediction of microRNA Targets by Ranks Aggregation. Front Genet 2020;10:1330. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Bronisz A, Rooj AK, Krawczyński K, et al. The nuclear DICER-circular RNA complex drives the deregulation of the glioblastoma cell microRNAome. Sci Adv 2020;6:eabc0221. [Crossref] [PubMed]
- Zhu J, Ye J, Zhang L, et al. Differential Expression of Circular RNAs in Glioblastoma Multiforme and Its Correlation with Prognosis. Transl Oncol 2017;10:271-9. [Crossref] [PubMed]
- Yuan Y, Jiaoming L, Xiang W, et al. Analyzing the interactions of mRNAs, miRNAs, lncRNAs and circRNAs to predict competing endogenous RNA networks in glioblastoma. J Neurooncol 2018;137:493-502. [Crossref] [PubMed]
- Liu B, Liu Y, Chen J, et al. CARP is a novel caspase recruitment domain containing pro-apoptotic protein. Biochem Biophys Res Commun 2002;293:1396-404. [Crossref] [PubMed]
- Lu F, Xue JX, Hu YC, et al. CARP is a potential tumor suppressor in gastric carcinoma and a single-nucleotide polymorphism in CARP gene might increase the risk of gastric carcinoma. PLoS One 2014;9:e97743. [Crossref] [PubMed]
- Shi Y, Chen J, Li Z, et al. C10ORF97 is a novel tumor-suppressor gene of non-small-cell lung cancer and a functional variant of this gene increases the risk of non-small-cell lung cancer. Oncogene 2011;30:4107-17. [Crossref] [PubMed]
- Swayne LA, Murphy NP, Asuri S, et al. Novel Variant in the ANK2 Membrane-Binding Domain Is Associated With Ankyrin-B Syndrome and Structural Heart Disease in a First Nations Population With a High Rate of Long QT Syndrome. Circ Cardiovasc Genet 2017;10:e001537. [Crossref] [PubMed]
- Sucharski HC, Dudley EK, Keith CBR, et al. Mechanisms and Alterations of Cardiac Ion Channels Leading to Disease: Role of Ankyrin-B in Cardiac Function. Biomolecules 2020;10:211. [Crossref] [PubMed]
- Watanabe H, Minamino T. Rare Variants in ANK2 Associated With Various Inherited Arrhythmia Syndromes. Circ J 2016;80:2423-4. [Crossref] [PubMed]
- Yanan F, Rui L, Xiaoying L, et al. Association between ACVR2A gene polymorphisms and risk of hypertensive disorders of pregnancy in the northern Chinese population. Placenta 2020;90:1-8. [Crossref] [PubMed]
- Glotov AS, Kazakov SV, Vashukova ES, et al. Targeted sequencing analysis of ACVR2A gene identifies novel risk variants associated with preeclampsia. J Matern Fetal Neonatal Med 2019;32:2790-6. [Crossref] [PubMed]
- Li G, Guo X. LncRNA STARD13-AS blocks lung squamous carcinoma cells growth and movement by targeting miR-1248/C3A. Pulm Pharmacol Ther 2020;64:101949. [Crossref] [PubMed]
- Qin WJ, Wang WP, Wang XB, et al. MiR-1290 targets CCNG2 to promote the metastasis of oral squamous cell carcinoma. Eur Rev Med Pharmacol Sci 2019;23:10332-42. [PubMed]
- Ma Q, Wang Y, Zhang H, et al. miR-1290 Contributes to Colorectal Cancer Cell Proliferation by Targeting INPP4B. Oncol Res 2018;26:1167-74. [Crossref] [PubMed]
- Ito H, Tsunoda T, Riku M, et al. Indispensable role of STIL in the regulation of cancer cell motility through the lamellipodial accumulation of ARHGEF7-PAK1 complex. Oncogene 2020;39:1931-43. [Crossref] [PubMed]
- Lei X, Deng L, Liu D, et al. ARHGEF7 promotes metastasis of colorectal adenocarcinoma by regulating the motility of cancer cells. Int J Oncol 2018;53:1980-96. [Crossref] [PubMed]
- Farlie P, Reid C, Wilcox S, et al. Ypel1: a novel nuclear protein that induces an epithelial-like morphology in fibroblasts. Genes Cells 2001;6:619-29. [Crossref] [PubMed]
- Shembade N, Parvatiyar K, Harhaj NS, et al. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J 2009;28:513-22. [Crossref] [PubMed]
- Shembade N, Harhaj EW. Elucidating dynamic protein-protein interactions and ubiquitination in NF-κB signaling pathways. Methods Mol Biol 2015;1280:283-95. [Crossref] [PubMed]
- Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 2007;13:460-9. [Crossref] [PubMed]
- Harris J, Olière S, Sharma S, et al. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J Immunol 2006;177:2527-35. [Crossref] [PubMed]
- Soubannier V, Stifani S. NF-κB Signalling in Glioblastoma. Biomedicines 2017;5:29. [Crossref] [PubMed]
- Min C, Eddy SF, Sherr DH, et al. NF-kappaB and epithelial to mesenchymal transition of cancer. J Cell Biochem 2008;104:733-44. [Crossref] [PubMed]
- Xie TX, Xia Z, Zhang N, et al. Constitutive NF-kappaB activity regulates the expression of VEGF and IL-8 and tumor angiogenesis of human glioblastoma. Oncol Rep 2010;23:725-32. [PubMed]
- Safa AR. Resistance to Cell Death and Its Modulation in Cancer Stem Cells. Crit Rev Oncog 2016;21:203-19. [Crossref] [PubMed]
- Li F, Zhou K, Gao L, et al. Radiation induces the generation of cancer stem cells: A novel mechanism for cancer radioresistance. Oncol Lett 2016;12:3059-65. [Crossref] [PubMed]
- Mattioni A, Castagnoli L, Santonico E. RNF11 at the Crossroads of Protein Ubiquitination. Biomolecules 2020;10:1538. [Crossref] [PubMed]
- Rao D, Guan S, Huang J, et al. miR-425-5p Acts as a Molecular Marker and Promoted Proliferation, Migration by Targeting RNF11 in Hepatocellular Carcinoma. Biomed Res Int 2020;2020:6530973. [Crossref] [PubMed]
- Mattioni A, Boldt K, Auciello G, et al. Ring Finger Protein 11 acts on ligand-activated EGFR via the direct interaction with the UIM region of ANKRD13 protein family. FEBS J 2020;287:3526-50. [Crossref] [PubMed]