
GLIPR1 regulates the TIMP1-CD63-ITGB1-AKT signaling pathway in glioma cells and induces malignant transformation of astroglioma
Introduction
Glioma (GM) is a highly aggressive malignant tumor in the central nervous system, accounting for up to 10–15% of all intracranial tumors (1). Compared with other nervous system tumors, GM has a higher tendency to metastasize, the overall prognosis is poor and the survival rate is lower (2). Astrocytoma (ACM) is a common tumor among GM, with specific morphological changes and biological behavior that is transformed from astrocytes, which account for more than 40% of GM (3-5). ACM grows slowly and cannot be completely removed by surgery, and its prognosis of patients is generally poor, especially for patients with highly malignant glioblastoma (6). Classical histopathological methods are very difficult to determine whether glial cells are tumors or reactive hyperplasia. In addition, the prognosis and response to treatment between different grades or the same grade of ACM vary greatly, and the classification methods widely used in clinical practice cannot fully reflect the true prognosis of patients. Therefore, it is necessary to find and discover new diagnostic and prognostic tumor markers related to ACM to improve the diagnosis rate, reduce the poor prognosis, and prolong the survival of patients.
Glioma pathogenesis-related protein 1 (GLIPR1), also known as related to testis-specific, vespid, and pathogenesis protein 1 (RTVP-1), is a membrane protein that belongs to the CAP family of cysteine-rich secretory proteins (7). Recent reports have indicated that GLIPR1 is closely associated with the development of tumors, especially GM (8,9). The high expression of GLIPR1 was found in GM cells but not in tumors or other cells of the central nervous system (10,11). Jacoby reported that GLIPR1 was highly expressed in glioblastoma and had carcinogenic characteristics (8). Ziv-Av et al. (9) found that GLIPR1 was highly expressed in GM and enhanced the invasion and migration of GM cells. Based on these observations, investigating the correlation between GLIPR1 and GM and exploring the specific mechanism by which GLIPR1 influences GM are expected to be promising for the prospective treatment of GM.
Accumulating evidence indicates that the TIMP1-CD63-ITGB1-AKT (tissue inhibitor of metalloproteinases 1, TIMP1; lysosomal membrane-associated glycoprotein 3, CD63; integrin beta 1, ITGB1; protein kinase B, AKT) cascade signaling pathway is closely related to the differentiation and migration of cancer cells, and this pathway may be regulated by GLIPR1. Through the analysis of biographical information (Starbase: https://starbase.sysu.edu.cn), it is found that GLIPR1 and CD63 have a positive regulatory relationship. In addition, studies have shown that the oncogenic function of GLIPR1 was mediated by the interaction of its cell surface receptor CD63 to induce intracellular signal transduction (12). GLIPR1 could promote hypermigration of cells via the CD63-ITGB1 signaling pathway (13). However, the activation of CD63 requires the participation of TIMP1 to interact with it and activate its expression (14,15). Studies have also indicated that ITGB1-AKT was closely related to the development of liver cancer (16). In addition, it has been reported that CD31 induced metastasis of liver cancer cells through the ITGB1-Akt signaling pathway (17). Also, TIMP1 could interact with CD63 and activate the Akt signaling pathway to confer apoptosis resistance to melanoma cells (15). This work prompted us to further explore the relationship between GLIPR1 and the TIMP1-CD63-ITGB1-AKT signaling pathway to regulate the proliferation and invasion of ACM cells.
In the current work, the effect of overexpression or interference with GLIPR1 expression on the activity, proliferation, migration, and invasion of ACM cells was studied. Besides, the preliminary exploration of the activation effect of GLIPR1 on the TIMP1-CD63-ITGB1-AKT cascade signaling pathway was carried out to explore the potential mechanism of GLIPR1 inducing ACM. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2413/rc).
Methods
Cell culture and treatment
The human astrocyte cell line SVG p12 and human ACM cell lines including SHC-44, SW1783 were purchased from the Stem Cell Bank (Chinese Academy of Sciences, Shanghai, China). Cells were cultured with DMEM medium containing 10% fetal bovine serum (FBS) (all from Gibco, Gaithersburg, MD, USA). Cells were cultured in a constant temperature incubator (37 ℃) containing 5% CO2. After incubation for 2 days, cells in good condition were utilized for subsequent assays. In addition, another control group was set up (anti-GLIPR1 group), that was, 6 nM of anti-GLIPR1 antibody (ab198215, Abcam, Shanghai, China) was added to SHC-44 and SW1783 cell lines, respectively. In addition, the anti-AKT group (5 nM) was set to investigate its role in the proliferation and invasion of cells.
Plasmid construction and cell infection
Both the overexpression plasmid ov-GLIPR1 and the interference plasmid sh-GLIPR1 were constructed by GenePharma Co., Ltd (Shanghai, China). Cells were seeded into a 15-cm petri dish with DMEM medium at a density of 5×105/mL for 24 h before transfection. A total of 0.8 µg plasmid DNA was mixed with 2.0 µL Lipofectamine® 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) and then was diluted by 50 µL Opti-MEM. The diluted DNA and transfection reagent were mixed at room temperature for 20 min. Next, the mixture was added to cells. After transfection for 6 h (37 ℃, 5% CO2). The original medium was removed and then the complete medium containing 10% FBS was added and incubated for 24 h. The washed cells were incubated in a fresh complete medium containing 10% FBS for 24 h.
CCK-8 assay
Cells of each group in the logarithmic phase were prepared into a cell suspension and seeded into a 96-well plate at a density of 104/mL. The DMEM medium containing 10% FBS medium in each well was removed at 24, 36, and 48 h after culture, and then 10 µL CCK-8 reagent was added (Beyotime, Beijing, China), and then cells were cultured in the original medium for 2 h and shaken for about 1 min to make the staining uniform. After that, the absorbance at 450 nm was calculated for cell viability.
Cell colony-forming assay
The cells of each group were seeded in a 6-well plate with a density of 104/mL, and the number of seeded cells per well was 200. The medium was changed every 3 days and the culture was terminated after 2 weeks of continuous culture. Next, the washed cells were fixed at room temperature (30 min) after adding 500 µL of 14% paraformaldehyde to each well. Subsequently, cells were washed and 500 µL of 0.1% crystal violet aqueous solution (Beyotime) was added to each well, and then cells were incubated at room temperature for 15 min. After staining, the cells were washed with double distilled water and the number of cloned cells formed was counted. A colony of ≥50 cells was regarded as a clone.
Transwell assay
The transwell chamber was placed in 24 well plates, then matrix glue was added to the transwell chamber, and the DMEM medium (complete) was added to the bottom chamber. The digested and resuspended cells were seeded in the upper chamber with a density of 3×104/chamber and then were cultured for 48 h. Subsequently, the medium was removed, cells were fixed with 4% polymethanol and stained with 0.1% crystal violet solution (Beyotime) for 20 min. The number of cells invading the basal chamber in each field was calculated under the microscope.
Cell scratch test
The digested cells of each group were seeded in 6-well plates (106/well) and cultured at 37 ℃, 5% CO2 and 100% relative humidity until the cells reached about 90% confluence. Then cells were scratched from top to bottom and the scratched cells were washed and then were continued to culture for 24 h under the same conditions and observed under the microscope, and the moving distance of cell front and scratch width were measured to analyze the cell migration capability.
Immunofluorescence
The immunofluorescence method was utilized for the visualization of protein expression and distribution in cells. Briefly, 4% formaldehyde was obtained for fixing cells, which were then permeated by 0.1% 100X Triton (Cayman Chemical, Ann Arbor, MI, USA). Cells were subsequently blocked with 10% goat serum (1 h) and incubated with primary antibody Anti-GLIPR1 (YB--13375R, 1:500, Ybscience, Shanghai, China) and Anti-CD63 (ab271286, 1:500, Abcam) at 4 ℃ overnight. The cells were washed with phosphate buffered saline (PBS) and incubated with fluorescent-labeled secondary antibody correspondingly. The results were observed and examined using a fluorescence microscope.
Western blot
Western blot was carried out for the measurement of protein levels. Briefly, the protein of each group of cells after transfection was extracted. The collected proteins were preliminarily isolated by SDS-PAGE and applied on a PVDF membrane. Next, they were blocked with 10% skimmed milk powder (4 ℃, 1 h) and then were in TBST buffer containing 10% skimmed milk powder and primary antibodies overnight. The primary antibodies included Anti-GLIPR1 (ab198215, 1:1,000), Anti-TIMP1 (ab211926, 1:1,000), Anti-CD63 (ab271286, 1:1,000), Anti-ITGB1 (ab30394, 1:1,000), Anti-AKT (ab179463, 1:1,000), and Anti-β-actin (ab8226, 1:1,000). The corresponding secondary antibodies were applied together with enhanced chemiluminescence (ECL) chromogenic solution. The density of protein bands was examined by using the image analyzer quantitative system. All primary antibodies were purchased from Abcam.
Co-IP assay
The Co-IP assay was performed according to the instructions of the Co-IP kit (Thermo Fisher Scientific, Los Angeles, CA, USA). Briefly, the cell lysate was added to each well (150 µL/well) of the six-well plate containing cells and shaken for 30 min on ice. After lysis, they were centrifuged (12,000 rpm) at 4 ℃ for 15 min. The protein supernatant was collected, and a little lysate was taken for subsequent detection of the expression of each protein. The remaining lysate was added to the GLIPR1 or CD63 antibody and incubated overnight at 4 ℃ with shaking. Protein A + G (20 µL) pretreated with lysis buffer was added to the cell lysate incubated with each antibody overnight and incubated at 4 ℃ for 2–4 h. After immunoprecipitation, they were centrifuged (3,000 rpm) at 4 ℃ for 3 min and the supernatant was removed. Next, SDS-PAGE was performed to detect protein expression.
Statistical analysis
At least three independent repeats were evaluated for each experiment. All statistical data were presented as the mean ± SD. All tests were performed by using SPSS 20.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 8.0 with the Student’s t-test. P<0.05 was regarded as significant.
Ethical statement
The research protocol was reviewed and approved by the Ethics Committee and Institutional Review Board of the Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (No. 20190302).
Results
GLIPR1 promoted the viability of ACM cells
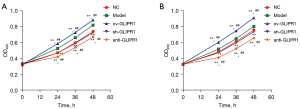
CCK-8 assay was employed to evaluate the effect of overexpression or interference with GLIPR1 on the viability of SHC-44 and SW1783 cells. Figure 1A shows that compared with the Model or normal (NC) group, the viability of SHC-44 cells in the ov-GLIPR1 group was greatly elevated at 24, 36, and 48 h, while those in the sh-GLIPR1 group or anti-GLIPR1 group was significantly reduced. Similarly, a similar trend could also be observed in SW1783 cells (Figure 1B). The results indicate that GLIPR1 can affect the viability of ACM cells.
GLIPR1 induced the proliferation of ACM cells
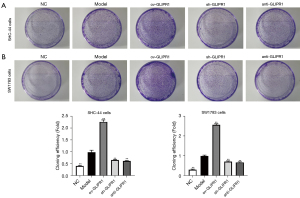
To analyze the influence of GLIPR1 on the proliferation of ACM cells, clone formation experiments were performed (Figure 2A,2B). The results showed that in SHC-44 and SW1783 cells, the cloning efficiency of cells transfected with ov-GLIPR1 was remarkably higher than that in the Model group, while the cloning efficiency of cells in the sh-GLIPR1 and anti-GLIPR1 groups was significantly reduced, which suggests that the GLIPR1 expression can promote the proliferation of SHC-44 and SW1783 cells.
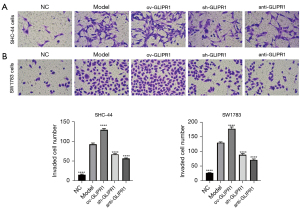
GLIPR1 enhanced the invasion of ACM cells
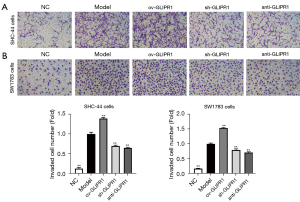
The invasion capability of ACM cells was tested by the transwell experiment (Figure 3A,3B). The results showed that in SHC-44 and SW1783 cells, the number of cells invading the lower chamber in the ov-GLIPR1 group was greatly higher than that in the Model group, while the number of cells invading the lower chamber in the sh-GLIPR1 and anti-GLIPR1 groups was greatly lower than Model group. Our suggests that the GLIPR1 expression can increase the invasion capability of SHC-44 and SW1783 cells.
GLIPR1 improved the migration of ACM cells
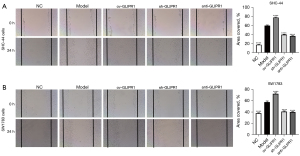
The cell scratch test was employed to evaluate the role of GLIPR1 in ACM cell migration. When the cells grow to about 90% confluence, a scratch of cells was made and observed (0 h), and then the cells were cultured for 24 h. In SHC-44 (Figure 4A) and SW1783 (Figure 4B) cells, the scratched coverage area of the ov-GLIPR1 group was markedly larger than that of the Model group, while the scratched coverage area of the sh-GLIPR1 and anti-GLIPR1 groups was greatly smaller than Model group. Accordingly, overexpression of GLIPR1 also significantly increased the migration of SHC-44 (Figure 5A) and SW1783 (Figure 5B) cells after scratching. Our results show that the GLIPR1 expression can improve the migration capability of ACM cells.
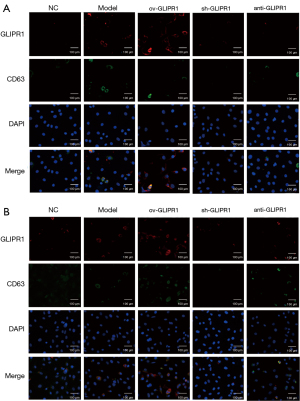
GLIPR1 regulated the CD63 expression
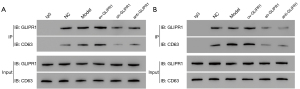
To explore the effect of GLIPR1 on CD63 expression, immunofluorescence assays were carried out. The results (Figure 6A,6B) show that GLIPR1 and CD63 had co-localization on the cell membrane and in the cytoplasm of the two cells (SHC-44 and SW1783). Accordingly, the Co-IP assay also illustrates GLIPR1 and CD63 had co-localization (Figure 7). This result confirms that they had an interaction. In addition, in the Model group and ov-GLIPR1 group, this phenomenon was particularly obvious, which suggested that GLIPR1 might regulate the CD63 expression.
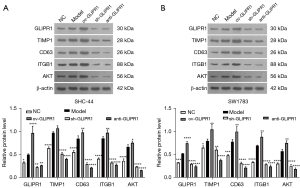
GLIPR1 activated the TIMP1-CD63-ITGB1-AKT signaling pathway
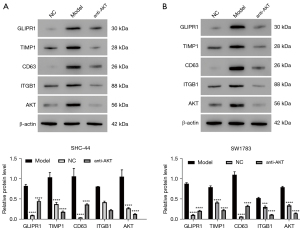
We used western blot assay to analyze the expression levels of GLIPR1, TIMP1, CD63, ITGB1, and AKT proteins in SHC-44 and SW1783 cells. As can be seen from Figure 8A,8B, the protein expressions of GLIPR1, TIMP1, CD63, ITGB1, and AKT in the ov-GLIPR1 group were greatly elevated compared with the MC group. In contrast, their expression levels of sh-GLIPR1 and anti-GLIPR1 groups were significantly reduced. Moreover, anti-AKT treatment significantly decreased the protein expression levels of GLIPR1, TIMP1, CD63, ITGB1, and AKT (Figure 9A,9B). Our results indicate that the GLIPR1 expression activates the TIMP1-CD63-ITGB1-AKT signaling pathway.
Discussion
ACM is the most common neuroepithelial tumor (18). Existing treatment methods, including conventional treatment, molecular targeted therapy, and drug therapy, have improved the treatment effect of the disease than before but are still limited by the drug resistance mechanism and the recurrence-prone characteristics of tumors. The overall prognosis of ACM patients is still poor, especially for malignant ACM, its incidence rate has been increasing year in recent years, and patients die within one and a half years after diagnosis (19). Therefore, it is particularly important to investigate the molecular mechanism of tumor occurrence and find a new effective treatment target.
At present, the prognosis of GM, especially malignant GM, is still not optimistic. GLIPR1 was found early as a proto-oncogene in human GMs (10), and its high expression can promote the proliferation, survival, invasion, and metastasis of GM cells (20). In addition, it is reported that GLIPR1 is related to the malignant degree of GM, and it plays an important role in regulating the proliferation, survival, and infiltration of GM cells (21). The siRNA transfection experiment proved that knocking down the expression of GLIPR1 could significantly inhibit the proliferation of GM U87 and A172 cells and induce apoptosis (8), whereas the overexpression of GLIPR1 could significantly promote the proliferation, survival, and invasion of U87 and A172 cells, and inhibit cell apoptosis (8). At the same time, the overexpression of GLIPR1 in human neural stem cells could induce their transformation to a mesenchymal phenotype (8). The silence of GLIPR1 in GM stem cells weakened its conversion to mesenchymal phenotype and the stemness of the cells. Also, silencing GLIPR1 could increase the survival of tumor-bearing mice in xenograft GM stem cells. These works all indicate that GLIPR1 is involved in the regulation of the occurrence and development of GMs, and is a key factor in the study of the pathogenesis of GMs. Consistent with the above reports, our results reveal that GLIPR1 expression promotes the viability, promotion, invasion, and migration of SHC-44 and SW1783 cells, which may be partly explained by the promoter methylation level of GLIPR1. The degree of methylation of the GLIPR1 promoter is closely related to the expression of GLIPR1 (22,23). It is reported that the GLIPR1 promoter had a low degree of methylation in GM (23). Therefore, the methylation of the GLIPR1 promoter may be one of the important factors affecting the expression of GLIPR1 in ACM.
CD63 has been regarded as an important factor in the development of solid tumors, and many reports have indicated a link between CD63 expression and many types of tumors (24). Based on biological information (Starbase: https://starbase.sysu.edu.cn), we found that GLIPR1 and CD63 have a positive regulatory relationship, and our results also illustrate this point. Our current research results found that GLIPR1 could interact with CD63 to regulate the expression of CD63 in SHC-44 and SW1783 cells, this is the first reported in the literature so far. Similar results were also reported by Warner et al. (12). However, it should be noted that the activation of CD63 may require the participation of TIMP1. It has been reported that TIMP1 was identified as a prognostic marker of GM, which could interact with CD63 in ACM cells and was significantly correlated with CD63 expression (25). The activated CD63 could further activate its ligand ITGB1 to enhance cell invasion (13,26). Studies have shown that ITGB1 might be related to the metastasis of tumor cells (27). AKT was the target protein of ITGB1, overexpression of ITGB1 could increase the expression of AKT and thus induce the growth of ACM cells (28), and this similar result has also been observed in other cancer models (17). Consistent with these reports, we found that GLIPR1 significantly activated the TIMP1-CD63-ITGB1-AKT signaling pathway, thereby enhancing the proliferation and invasion of ACM cells. These studies suggest that activation of the TIMP1-CD63-ITGB1-AKT pathway may play an important role in the proliferation and invasion of tumor cells. However, the relevant mechanism is still unknown, and more in-depth investigations are needed in the future.
Together, GLIPR1 is a key factor leading to the proliferation and metastasis of GM. It can improve the proliferation and invasion of ACM cells by activating the TIMP1-CD63-ITGB1-AKT signaling pathway, and it can be an important target for the prevention or management of ACM in the clinic.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2413/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2413/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2413/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research protocol was reviewed and approved by the Ethics Committee and Institutional Review Board of the Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (No. 20190302).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jiang T, Mao Y, Ma W, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett 2016;375:263-73. [Crossref] [PubMed]
- Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev 2017;40:1-14. [Crossref] [PubMed]
- Walker DG, Kaye AH. Diagnosis and management of astrocytomas, oligodendrogliomas and mixed gliomas: a review. Australas Radiol 2001;45:472-82. [Crossref] [PubMed]
- Mathieu D, Fortin D. The role of chemotherapy in the treatment of malignant astrocytomas. Can J Neurol Sci 2006;33:127-40. [Crossref] [PubMed]
- Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol 2004;108:49-56. [Crossref] [PubMed]
- Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007;21:2683-710. [Crossref] [PubMed]
- Asojo OA, Koski RA, Bonafé N. Structural studies of human glioma pathogenesis-related protein 1. Acta Crystallogr D Biol Crystallogr 2011;67:847-55. [Crossref] [PubMed]
- Jacoby E, Yalon M, Leitner M, et al. Related to testes-specific, vespid and pathogenesis protein-1 is regulated by methylation in glioblastoma. Oncol Lett 2014;7:1209-12. [Crossref] [PubMed]
- Ziv-Av A, Giladi ND, Lee HK, et al. RTVP-1 regulates glioma cell migration and invasion via interaction with N-WASP and hnRNPK. Oncotarget 2015;6:19826-40. [Crossref] [PubMed]
- Murphy EV, Zhang Y, Zhu W, et al. The human glioma pathogenesis-related protein is structurally related to plant pathogenesis-related proteins and its gene is expressed specifically in brain tumors. Gene 1995;159:131-5. [Crossref] [PubMed]
- Rich T, Chen P, Furman F, et al. RTVP-1, a novel human gene with sequence similarity to genes of diverse species, is expressed in tumor cell lines of glial but not neuronal origin. Gene 1996;180:125-30. [Crossref] [PubMed]
- Warner RB, Najy AJ, Jung YS, et al. Establishment of Structure-Function Relationship of Tissue Inhibitor of Metalloproteinase-1 for Its Interaction with CD63: Implication for Cancer Therapy. Sci Rep 2020;10:2099. [Crossref] [PubMed]
- Ólafsson EB, Ross EC, Varas-Godoy M, et al. TIMP-1 promotes hypermigration of Toxoplasma-infected primary dendritic cells via CD63-ITGB1-FAK signaling. J Cell Sci 2019;132:jcs225193. Erratum in: J Cell Sci 2019;132:jcs230920. [Crossref] [PubMed]
- Dong J, Ma Q. TIMP1 promotes multi-walled carbon nanotube-induced lung fibrosis by stimulating fibroblast activation and proliferation. Nanotoxicology 2017;11:41-51. [Crossref] [PubMed]
- Toricelli M, Melo FH, Peres GB, et al. Timp1 interacts with beta-1 integrin and CD63 along melanoma genesis and confers anoikis resistance by activating PI3-K signaling pathway independently of Akt phosphorylation. Mol Cancer 2013;12:22. [Crossref] [PubMed]
- Guo D, Zhang D, Ren M, et al. THBS4 promotes HCC progression by regulating ITGB1 via FAK/PI3K/AKT pathway. FASEB J 2020;34:10668-81. [Crossref] [PubMed]
- Zhang YY, Kong LQ, Zhu XD, et al. CD31 regulates metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma via the ITGB1-FAK-Akt signaling pathway. Cancer Lett 2018;429:29-40. [Crossref] [PubMed]
- Marumoto T, Saya H. Molecular biology of glioma. Adv Exp Med Biol 2012;746:2-11. [Crossref] [PubMed]
- Mrugala MM. Advances and challenges in the treatment of glioblastoma: a clinician's perspective. Discov Med 2013;15:221-30. [PubMed]
- Rosenzweig T, Ziv-Av A, Xiang C, et al. Related to testes-specific, vespid, and pathogenesis protein-1 (RTVP-1) is overexpressed in gliomas and regulates the growth, survival, and invasion of glioma cells. Cancer Res 2006;66:4139-48. [Crossref] [PubMed]
- Caplen NJ, Fleenor J, Fire A, et al. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 2000;252:95-105. [Crossref] [PubMed]
- Karantanos T, Tanimoto R, Edamura K, et al. Systemic GLIPR1-ΔTM protein as a novel therapeutic approach for prostate cancer. Int J Cancer 2014;134:2003-13. [Crossref] [PubMed]
- Giladi ND, Ziv-Av A, Lee HK, et al. RTVP-1 promotes mesenchymal transformation of glioma via a STAT-3/IL-6-dependent positive feedback loop. Oncotarget 2015;6:22680-97. [Crossref] [PubMed]
- Koh HM, Jang BG, Kim DC. Prognostic Value of CD63 Expression in Solid Tumors: A Meta-analysis of the Literature. In Vivo 2020;34:2209-15. [Crossref] [PubMed]
- Aaberg-Jessen C, Sørensen MD, Matos ALSA, et al. Co-expression of TIMP-1 and its cell surface binding partner CD63 in glioblastomas. BMC Cancer 2018;18:270. [Crossref] [PubMed]
- Ólafsson EB, Barragan A. The unicellular eukaryotic parasite Toxoplasma gondii hijacks the migration machinery of mononuclear phagocytes to promote its dissemination. Biol Cell 2020;112:239-50. [Crossref] [PubMed]
- Yi Y, Fang Y, Wu K, et al. Comprehensive gene and pathway analysis of cervical cancer progression. Oncol Lett 2020;19:3316-32. [Crossref] [PubMed]
- Wang J, Cai C, Nie D, et al. FRK suppresses human glioma growth by inhibiting ITGB1/FAK signaling. Biochem Biophys Res Commun 2019;517:588-95. [Crossref] [PubMed]


