
The identification of hub biomarkers and pathways in lung cancer and prognostic evaluation
Introduction
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer death in 2020, which account for approximately one-tenth cancers diagnosed and one in 5 deaths. There are estimated 2.2 million new cancer cases and 1.8 million deaths in the world (1). Lung cancer can be divided into two histologic subtypes, including non-small cell lung cancer (NSCLC, about 85% of all lung cancer) and small cell lung cancer (SCLC, about 15%). NSCLC is made up of three major histologic subtypes: large-cell lung, two major pathological types adenocarcinoma (LUAD) and lung squamous-cell carcinoma (LUSC) (2). The treatments of lung cancer mainly included surgery, chemotherapy, radiotherapy and immunotherapy. As we know, there are different treatment options and prognosis for these subtypes. Therapeutic advances contributed to survival gains. With the progress on targeted therapies and immunotherapies, the 2-years survival rate for NSCLC increased from 34% during 2009–2010 to 42% during 2015–2016 in the United States, but SCLC survival remained low and steady at 14% to 15% (3-5). There are significant differences in survival between NSCLC and SCLC. Therefore, it is important to assess the difference of lung cancer to detect prognostic markers which are likely to affect future treatments and prognosis.
Gene expression profile array and bioinformatics analysis have been applied to study potential clinical biomarkers and molecular mechanisms. Some key differentially expressed genes (DEGs) have been identified by integrated bioinformatics analysis and are significantly associated with the treatments and prognosis in some cancers (6-11). Relevant biomarkers have been used as valuable tools in the prognosis and prediction of therapy response, significantly influencing the clinical course and outcome of the disease (12). In our study, we selected gene expression profile from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and analyzed the differential expression genes between differential lung cancer tissues and normal tissues to explore the hub pathways and key genes. We applied integrated bioinformatics methods to further investigate potential gene biomarkers and molecular mechanisms in lung cancer. Novel prognostic biomarkers will further inform clinical therapeutic decision-making. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-245/rc).
Methods
Microarray data and identification of DEGs
The gene expression profiles of lung cancer (GSE43346, GSE40275 and GSE18842) were downloaded from Gene Expression Omnibus public database (13-16). The above three gene expression profiles were performed by the Affymetrix Human Gene Expression Array. GSE43346 contains 42 normal tissue samples and 23 SCLC samples. GSE40275 includes 43 normal tissue samples, 19 SCLC samples and 16 NSCLC samples. GSE18842 includes 45 normal tissue samples and 46 NSCLC samples.
GEO2R online tool and Venn diagram software were applied to screen overlap DEGs from above three gene expression profiles. |logFC| >2 and adjusted P value <0.05 were used as cutoff criteria by GEO2R online tools. The logFC <−2 or logFC >2 were considered down-regulated or up-regulated genes, respectively.
Gene Ontology (GO) and Kyoto Encyclopedia of Gene and Genome (KEGG) functional enrichment analysis
The GO and KEGG were performed using Database for Annotation, Visualization and Integrated Discovery v6.8 (DAVID, https://david.ncifcrf.gov/) (17,18). The DAVID v6.8, an online set of functional annotation tools, was used to analyze biological process, cellular component, molecular function and pathways for DEGs. GO terms and KEGG pathways with P value <0.05 were considered statistical significant.
Protein-protein interaction network construction and module analysis
Protein-protein interaction network was obtained through the Search Tool for the Retrieval of Interacting Genes database (STRING, http://string-db.org) (19). The plugin MCODE of Cytoscape was applied to detect significant modules in the protein-protein interaction network (https://cytoscape.org/). The cutoff criteria were set with degree cutoff =2, node score cutoff =0.2, maximum depth =100, and k-core =2 (20). The interactions of module DEGs in the PPI networks were analysed using plugin cytoHubba (21). The specific connectivity genes that overlapped in the NSCLC, SCLC or NSCLC & SCLC PPI networks, are defined as hub DEGs.
Validation of hub DEGs and survival analysis
The online Kaplan-Meier plotter database (https://kmplot.com/analysis/) and The GEPIA server (http://gepia.cancer-pku.cn/) were applied to assess the survival rate of patients with LUAD or LUSC (22,23). The Kaplan-Meier plotter database, which the tool of a meta-analysis-based discovery and validation of survival biomarkers, includes 54,000 genes on survival in 21 cancer types including 3,452 lung cancer patients. The criteria we selected were HR with 95% CI and log-rank P<0.05 as a threshold.
The GEPIA is an interactive web serve of analyzing tumor/normal differential expression analysis, correlation analysis and patient survival analysis. The GEPIA was used to get stage plots and further validate the expression of hub DEGs between LUAD, LUSC and normal lung tissues (P<0.05).
Ethical consideration
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Differentially expressed genes in SCLC and NSCLC
We extracted 1,945 and 1,137 DEGs using the online GEO2R tool between SCLC and normal lung tissues from GSE43346 and GSE40275. The 1,014 and 1,367 DEGs were extracted from GSE18842 and GSE40275 in NSCLC tissues compared with normal tissues. We identified 84 overlap DEGs in SCLC & NSCLC tissues by online Venn diagram software, including 54 up-regulated and 30 down-regulated DEGs. The 70 up-regulated and 17 down-regulated DEGs were identified unique to SCLC. The 20 up-regulated and 8 down-regulated DEGs were identified unique to NSCLC (Table 1 and Figure 1).
Table 1
| Cancer | DEGs | No. | Genes name |
|---|---|---|---|
| SCLC & NSCLC | Up-regulated | 54 | TPX2, CCNB1, HMGB3, DSP, GINS1, ANLN, UCHL1, EZH2, CHEK1, KIF11, CDC6, AURKA, KIF14, KIF4A, TYMS, CDCA7, MELK, NDC80, RFC4, CCNA2, BUB1, PBK, NUF2, PRR11, PTTG1, MMP12, UBE2T, ECT2, KIF23, DEPDC1, GGH, ASPM, ATAD2, BRIP1, UBE2C, CCNB2, PRC1, CEP55, RRM2, TOP2A, HELLS, CCNE2, BUB1B, RAD51AP1, MKI67, DTL, EXO1, KIF20A, KIAA0101, TTK, CDKN3, NCAPG, CENPF, NUSAP1 |
| Down-regulated | 30 | CHRDL1, EGR1, FHL1, AOC3, ZFP36, ZBTB16, EDNRB, ARRB1, SH2D3C, TNS1, C7, GPM6A, GADD45B, MFAP4, PTGDS, SDPR, NR4A1, FOS, TPSB2, AQP1, ADH1B, FABP4, FAM107A, PGM5, GPX3, FXYD1, FOSB, VIPR1, CFD, FBLN5 | |
| Only SCLC | Up-regulated | 70 | DONSON, DEPDC1B, RAB3IP, PCSK1, RFC3, BRCA1, ACTL6A, ISL1, PLK1, NUP62CL, MYEF2, ASF1B, CDH2, KIFC1, CBFA2T2, ZNF711, TMPO, DCX, RNF182, FANCA, GDAP1, PCNA, TPH1, SLC36A4, LOC643201, GNG4, H2AFY2, CCDC14, MIAT, STMN1, BEST3, TPD52, INTS7, NOL4, STXBP5L, TUBB2B, CEP78, GPR137C, PGAP1, HOXD10, NELL1, RAB3B, PMAIP1, DDC, MSH2, RRM1, SCG3, ESCO2, KIF1A, CBX3, MEST, MPHOSPH9, NRCAM, CDKN2C, GRP, RIMS2, MCM6, AGPAT5, CENPI, SCN8A, FZD3, GMNN, SSX2IP, SLCO5A1, ASCL1, CDKN2A, RFC5, USP1, LOC81691, ST18 |
| Down-regulated | 17 | LAMA2, HLA-DRB4, DPP4, MAOA, C3, RBPMS, ADAMTS1, AQP3, CD74, SYNE1, ANPEP, CCL21, RNASE1, MYL9, SNTB1, KIAA1462, DCN | |
| Only NSCLC | Up-regulated | 20 | SERPINB5, AKR1B10, GJB6, ARNTL2, SLC7A11, SPRR1A, PLOD2, KRT6A, GCLC, FAP, FAM83B, NQO1, PSAT1, S100A2, SULF1, CLDN1, GPR87, CP, STEAP1, RPL39L |
| Down-regulated | 8 | GRK5, KLF2, ADARB1, CLDN5, DENND3, CCBE1, MFNG, SELENBP1 |
SCLC, small cell lung cancer; NSCLC, non-small cell lung cancer; DEGs, differentially expressed genes.
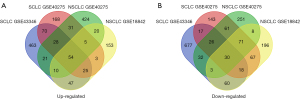
Functional and pathway enrichment analysis
The online DAVID was utilized for GO and KEGG enrichment analysis of overlap DEGs in lung cancer. The GO analysis includes biological processes, cellular components, and molecular functions. For biological process analysis, 54 up-regulated and 30 down-regulated DEGs in SCLC & NSCLC were mainly related to mitotic nuclear division, cell division, mitotic cytokinesis, regulation of cell cycle, DNA replication and mitotic spindle organization, while the DEGs unique to SCLC were mainly involved in DNA damage response, detection of DNA damage, DNA replication, neuron migration, error-prone translesion synthesis, error-free translesion synthesis and negative regulation of cyclin-dependent protein serine/threonine kinase activity. The biological process analysis showed DEGs unique to NSCLC were associated with oxidation-reduction process, morphogenesis of an epithelium, aging, calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules, positive regulation of vascular endothelial growth factor production and endothelial cell migration. Furthermore, cellular component analysis indicated that overlap DEGs in SCLC & NSCLC were located in the nucleus, nucleoplasm, midbody, spindle, cytoplasm and cytosol, while DEGs unique to SCLC in DNA replication factor C complex, chromatin, nuclear envelope, nuclear chromosome, telomeric region, transport vesicle membrane and nucleoplasm, and DEGs unique to NSCLC in extracellular space and extracellular exosome. Additionally, the results of molecular function analysis indicated that overlap DEGs in lung cancer were particularly enriched in protein binding, ATP binding, protein kinase binding, microtubule binding, chromatin binding and microtubule motor activity, while DEGs unique to SCLC in dinucleotide insertion or deletion binding, enzyme binding, mutLalpha complex binding, damaged DNA binding, DNA clamp loader activity and single-stranded DNA-dependent ATPase activity, and DEGs unique to NSCLC in structural molecule activity (Table 2).
Table 2
| Cancer | Category | Term | Count | % | P value | FDR |
|---|---|---|---|---|---|---|
| SCLC & NSCLC | GOTERM_BP_DIRECT | GO:0007067~mitotic nuclear division | 17 | 20.24 | 5.53E-14 | 2.09E-11 |
| GOTERM_BP_DIRECT | GO:0051301~cell division | 19 | 22.62 | 6.21E-14 | 2.09E-11 | |
| GOTERM_BP_DIRECT | GO:0000281~mitotic cytokinesis | 6 | 7.14 | 2.66E-07 | 5.97E-05 | |
| GOTERM_BP_DIRECT | GO:0051726~regulation of cell cycle | 8 | 9.52 | 2.43E-06 | 4.09E-04 | |
| GOTERM_BP_DIRECT | GO:0006260~DNA replication | 8 | 9.52 | 1.06E-05 | 0.001430782 | |
| GOTERM_BP_DIRECT | GO:0007052~mitotic spindle organization | 5 | 5.95 | 1.31E-05 | 0.001476073 | |
| GOTERM_CC_DIRECT | GO0005634~nucleus | 54 | 64.29 | 1.21E-10 | 1.78E-08 | |
| GOTERM_CC_DIRECT | GO0005654~nucleoplasm | 36 | 42.86 | 3.51E-09 | 2.58E-07 | |
| GOTERM_CC_DIRECT | GO0030496~midbody | 10 | 11.90 | 7.12E-09 | 3.49E-07 | |
| GOTERM_CC_DIRECT | GO0005819~spindle | 9 | 10.71 | 7.78E-08 | 2.86E-06 | |
| GOTERM_CC_DIRECT | GO0005737~cytoplasm | 45 | 53.57 | 2.61E-06 | 7.67E-05 | |
| GOTERM_CC_DIRECT | GO0005829~cytosol | 34 | 40.48 | 3.40E-06 | 8.33E-05 | |
| GOTERM_MF_DIRECT | GO:0005515~protein binding | 64 | 76.19 | 1.20E-07 | 2.35E-05 | |
| GOTERM_MF_DIRECT | GO:0005524~ATP binding | 22 | 26.19 | 2.67E-06 | 2.61E-04 | |
| GOTERM_MF_DIRECT | GO:0019901~protein kinase binding | 10 | 11.90 | 5.77E-05 | 0.003748425 | |
| GOTERM_MF_DIRECT | GO:0008017~microtubule binding | 7 | 8.33 | 4.00E-04 | 0.016613263 | |
| GOTERM_MF_DIRECT | GO:0003682~chromatin binding | 9 | 10.71 | 4.39E-04 | 0.016613263 | |
| GOTERM_MF_DIRECT | GO:0003777~microtubule motor activity | 5 | 5.95 | 5.11E-04 | 0.016613263 | |
| Only SCLC | GOTERM_BP_DIRECT | GO:0042769~DNA damage response, detection of DNA damage | 4 | 4.60 | 6.88E-04 | 0.34328918 |
| GOTERM_BP_DIRECT | GO:0006260~DNA replication | 6 | 6.90 | 9.15E-04 | 0.34328918 | |
| GOTERM_BP_DIRECT | GO:0001764~neuron migration | 5 | 5.75 | 0.001660073 | 0.415018297 | |
| GOTERM_BP_DIRECT | GO:0042276~error-prone translesion synthesis | 3 | 3.45 | 0.003726164 | 0.558924545 | |
| GOTERM_BP_DIRECT | GO:0070987~error-free translesion synthesis | 3 | 3.45 | 0.003726164 | 0.558924545 | |
| GOTERM_BP_DIRECT | GO:0045736~negative regulation of cyclin-dependent protein serine/threonine kinase activity | 3 | 3.45 | 0.004986668 | 0.621663134 | |
| GOTERM_CC_DIRECT | GO:0005663~DNA replication factor C complex | 3 | 3.45 | 3.11E-04 | 0.046840782 | |
| GOTERM_CC_DIRECT | GO:0000785~chromatin | 5 | 5.75 | 7.61E-04 | 0.046840782 | |
| GOTERM_CC_DIRECT | GO:0005635~nuclear envelope | 6 | 6.90 | 8.41E-04 | 0.046840782 | |
| GOTERM_CC_DIRECT | GO:0000784~nuclear chromosome, telomeric region | 5 | 5.75 | 0.003071132 | 0.12821977 | |
| GOTERM_CC_DIRECT | GO:0030658~transport vesicle membrane | 3 | 3.45 | 0.013254179 | 0.376099937 | |
| GOTERM_CC_DIRECT | GO:0005654~nucleoplasm | 22 | 25.29 | 0.013512573 | 0.376099937 | |
| GOTERM_MF_DIRECT | GO:0019899~enzyme binding | 6 | 6.90 | 0.019926207 | 1 | |
| GOTERM_MF_DIRECT | GO:0032405~MutLalpha complex binding | 2 | 2.30 | 0.0277565 | 1 | |
| GOTERM_MF_DIRECT | GO:0003684~damaged DNA binding | 3 | 3.45 | 0.035139311 | 1 | |
| GOTERM_MF_DIRECT | GO:0003689~DNA clamp loader activity | 2 | 2.30 | 0.036838541 | 1 | |
| GOTERM_MF_DIRECT | GO:0043142~single-stranded DNA-dependent ATPase activity | 2 | 2.30 | 0.045836807 | 1 | |
| Only NSCLC | GOTERM_BP_DIRECT | GO:0055114~oxidation-reduction process | 5 | 17.86 | 0.01408762 | 1 |
| GOTERM_BP_DIRECT | GO:0002009~morphogenesis of an epithelium | 2 | 7.14 | 0.022285495 | 1 | |
| GOTERM_BP_DIRECT | GO:0007568~aging | 3 | 10.71 | 0.028682612 | 1 | |
| GOTERM_BP_DIRECT | GO:0016338~calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | 2 | 7.14 | 0.033248128 | 1 | |
| GOTERM_BP_DIRECT | GO:0010575~positive regulation of vascular endothelial growth factor production | 2 | 7.14 | 0.042550374 | 1 | |
| GOTERM_BP_DIRECT | GO:0043542~endothelial cell migration | 2 | 7.14 | 0.045631924 | 1 | |
| GOTERM_CC_DIRECT | GO:0005615~extracellular space | 7 | 25.00 | 0.010282349 | 0.575811533 | |
| GOTERM_CC_DIRECT | GO:0070062~extracellular exosome | 9 | 32.14 | 0.037100315 | 1 | |
| GOTERM_MF_DIRECT | GO:0005198~structural molecule activity | 4 | 14.29 | 0.006274785 | 0.489433259 |
SCLC, small cell lung cancer; NSCLC, non-small cell lung cancer; FDR, false discovery rate.
The top 6 KEGG analysis results using DAVID software are shown in Table 3. The overlap DEGs in SCLC&NSCLC are mainly associated with cell cycle, the p53 signaling pathway, oocyte meiosis, progesterone-mediated oocyte maturation and HTLV-I infection pathways, and DEGs unique to SCLC enriched in mismatch repair, DNA replication, cell cycle, tryptophan metabolism, serotonergic synapse and nucleotide excision repair pathways. The DEGs unique to NSCLC were not enriched in any signaling pathway (all pathways P value >0.05).
Table 3
| Cancer | Category | Term | Count | % | P value | FDR |
|---|---|---|---|---|---|---|
| SCLC & NSCLC | KEGG_PATHWAY | hsa04110: cell cycle | 11 | 13.10 | 1.76E-09 | 1.46E-07 |
| KEGG_PATHWAY | hsa04115: p53 signaling pathway | 6 | 7.14 | 4.31E-05 | 0.001786741 | |
| KEGG_PATHWAY | hsa04114: oocyte meiosis | 6 | 7.14 | 4.71E-04 | 0.013041937 | |
| KEGG_PATHWAY | hsa04914: progesterone-mediated oocyte maturation | 4 | 4.76 | 0.014735684 | 0.277141512 | |
| KEGG_PATHWAY | hsa05166: HTLV-I infection | 6 | 7.14 | 0.016695272 | 0.277141512 | |
| Only SCLC | KEGG_PATHWAY | hsa03430: mismatch repair | 4 | 4.60 | 2.76E-04 | 0.029521615 |
| KEGG_PATHWAY | hsa03030: DNA replication | 4 | 4.60 | 0.001057086 | 0.056554103 | |
| KEGG_PATHWAY | hsa04110: cell cycle | 5 | 5.75 | 0.00508098 | 0.181221637 | |
| KEGG_PATHWAY | hsa00380: tryptophan metabolism | 3 | 3.45 | 0.021332245 | 0.514320928 | |
| KEGG_PATHWAY | hsa04726: serotonergic synapse | 4 | 4.60 | 0.024529657 | 0.514320928 | |
| KEGG_PATHWAY | hsa03420: nucleotide excision repair | 3 | 3.45 | 0.028840426 | 0.514320928 | |
| Only NSCLC | None |
KEGG, Kyoto Encyclopedia of Genes and Genome; SCLC, small cell lung cancer; NSCLC, non-small cell lung cancer; FDR, false discovery rate.
Protein-protein interaction network and module analysis
The overlap DEGs in SCLC&NSCLC, unique to SCLC or NSCLC were used to construct the protein-protein interaction network using STRING and Cytoscape, respectively. A total of 84 DEGs in SCLC&NSCLC were imported into online STRING, which contained 84 nodes and 1,122 edges. The 2 important modules were identified using Cytoscape MCODE, which contains 47 hub genes and 5 hub genes, respectively (Figure 2). The 7 of 12 topological analysis methods using cytoHubba have identified 100% (47/47) hub DEGs that we have screened using plugin MCODE in the SCLC & NSCLC PPI networks. The remaining 5 methods have identified at least 25 of the 47 hub DEGs (Table S1).
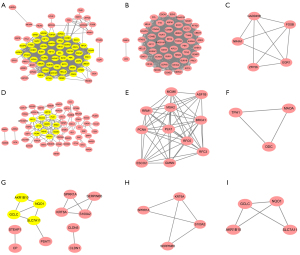
The 87 DEGs unique to SCLC were analyzed using STRING. The results of protein-protein interaction network showed that a total of 84 nodes and 123 edges were acquired. We applied Cytotype MCODE for further analysis to get two hub modules including 11 hub nodes and 3 hub nodes. The 7 of 12 methods using cytoHubba have identified at least 8 of the 11 hub DEGs we have screened using plugin MCODE unique to SCLC PPI networks (Table S2). We also imported 28 DEGs unique to NSCLC into online STRING for protein-protein interaction network. The 28 nodes and 13 edges were included in protein-protein interaction network. Two modules were obtained using Cytotype MCODE. Every module contained 4 hub nodes (Figure 2). The 11 of 12 methods using cytoHubba also have identified at least 7 of the 8 hub DEGs unique to NSCLC PPI networks (Table S3).
Survival analysis and cross-validation of hub DEGs
We used the Kaplan-Meier plotter database and GEPIA to further analyze prognosis value of hub DEGs in lung cancer. As we know, 85% of lung cancer patients were NSCLC, which mainly contains LUAD and LUSC. There was survival information of unique to NSLSC patients in above two databases. There are 1,925 NSCLC patients for survival analysis using Kaplan-Meier plotter database. We conducted cross-validation survival analysis of 52 overlap hub DEGs in SCLC & NSCLC and 8 hub DEGs unique to NSCLC associated with NSCLC patients. The results demonstrated that 12 overlap hub DEGs in SCLC & NSCLC and 2 hub DEGs unique to NSCLC were significantly associated with the overall survival of NSCLC patients (P<0.05, Table 4, Figures 3,4).
Table 4
| Cancer | Genes |
|---|---|
| SCLC & NSCLC | ANLN, CHEK1, DTL, ECT2, KIF11, MKI67, NCAPG, PRC1, PTTG1, RRM2, TYMS, KIF14 |
| Only NSCLC | KRT6A, SERPINB5 |
The overlap 12 hub DEGs in SCLC & NSCLC and 2 hub DEGs unique to in NSCLC were significantly associated with overall survival of NSCLC patients (P<0.05). DEGs, differentially expressed genes; GEPIA, Gene Expression Profiling Interactive Analysis; SCLC, small cell lung cancer; NSCLC, non-small cell lung cancer.
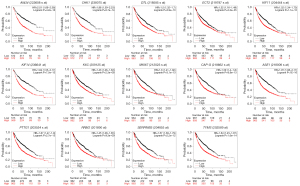
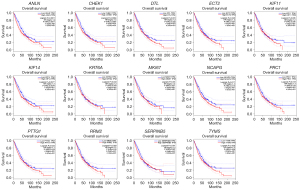
The online GEPIA software was used to validate the expression of 14 hub DEGs in NSCLC tissues compared with normal lung tissues. A total of 11 of 14 hub DEGs were also overexpressed in LAUD and LUSC (P<0.05), but the other 3 hub DEGs KIF14, KRT6A and SERPINB5 were only significantly different in LUSC not in LAUD (P>0.05) (Figure 5).
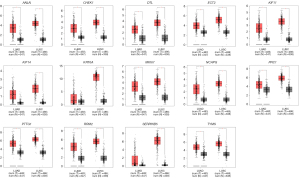
Pathway enrichment re-analysis and stage analysis of hub DEGs
The 14 hub DEGs were re-analyzed to identify the more important pathways using DAVID software. The results showed that the p53 signaling pathway and pyrimidine metabolism pathway were significantly associated with the survival of NSCLC patients (P<0.05). The RRM2, CHEK1 and SERPINB5 enriched in the p53 signaling pathway, RRM2 and TYMS enriched in pyrimidine metabolism pathway maybe play a key role in lung cancer. We used the GEPIA to validate the expression of these 4 hub DEGs in different stages of NSCLC. Statistical analysis identified the expression of RRM2, CHEK1, TYMS and SERPINB5 were significant differential across different stages (Table 5 and Figure 6).
Table 5
| Pathway ID | Name | Count | % | P value | Genes |
|---|---|---|---|---|---|
| hsa04115 | p53 signaling pathway | 3 | 21.43 | 5.54E-04 | RRM2, CHEK1, SERPINB5 |
| hsa00240 | Pyrimidine metabolism pathway | 2 | 14.29 | 0.057460904 | RRM2, TYMS |
| hsa04110 | Cell cycle pathway | 2 | 14.29 | 0.070192126 | PTTG1, CHEK1 |
DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genome.

Discussion
According to WHO criteria for lung tumors classification and diagnosis, lung cancer is generally divided into two histologic subtypes SCLC and NSCLC, and NSCLC is the main histological subtype of lung cancer. In order to identify the genetic differences between SCLC and NSCLC, we separately extracted DEGs in SCLC or NSCLC through the GEO database. Our study suggested hub DEGs play an important role not only in SCLC but also in NSCLC. We also found that some genes were only significantly differential expressed in SCLC or NSCLC. These results revealed that there are consistent differences and similarities between SCLC and NSCLC.
We further analyzed gene functional enrichment and interaction of DEGs using online DAVID, STRING database and Cytoscape MCODE. The overlap 84 DEGs found in both SCLC & NSCLC were mainly related to cell cycle, the p53 signaling pathway, oocyte meiosis, progesterone-mediated oocyte maturation and HTLV-I infection pathways. The DEGs unique to SCLC were enriched in mismatch repair, DNA replication, cell cycle, tryptophan metabolism, serotonergic synapse and nucleotide excision repair pathways. The DEGs unique to NSCLC were not associated with anyone pathway. Due to the lack of prognosis information of SCLC patients, we only carried out survival analysis of hub DEGs related to NSCLC patients. The results showed 14 hub DEGs were significantly associated with the overall survival of LAUD and LUSC patients. KEGG pathway enrichment re-analysis and stage analysis revealed that RRM2, CHEK1, TYMS and SERPINB5 maybe new effective biomarkers in NSCLC prognosis, which were enriched in the p53 signaling pathway and pyrimidine metabolism pathway.
The ribonucleotide reductase regulatory subunit M2 (RRM2), one of two non-identical subunits for ribonucleotide reductase, catalyzes ribonucleotides to form deoxyribonucleotides. Transcription of RRM2 results in two isoforms that differ in the lengths of their N-termini. RRM2 maintains to support DNA synthesis and repair and overexpressed in colorectal cancer, breast cancer, and cervical cancer (24-26). The expression of RRM2 in primary oral squamous cell carcinoma (OSCC) was significantly increased compared with normal tissues, and its overexpression was significantly associated with pathological grade, proliferation and migration, and recurrence in OSCC (27). High expression of RRM2 was associated with an immunosuppressive tumor-immune microenvironment and contributed to immune escape in prostate cancer (28). Studies have demonstrated that overexpressed miR-20a dramatically suppresses NSCLC cell proliferation and migration by inhibiting RRM2-mediated PI3K/Akt signaling, while the expression of RRM2 was upregulated in NSCLC (29). The RRM2 overexpression, as an independent predictive factor of poor prognosis in patients with lung adenocarcinoma, was significantly associated with tumor stage and TNM classification and reduced the activation of p53 signaling pathway (30).
The protein encoded by checkpoint kinase 1 (CHEK1), belongs to the Ser/Thr protein kinase family, and mediates cell cycle arrest in response to DNA damage or the presence of unreplicated DNA. This protein also integrates signals from ATM and ATR that are associated with chromatin in meiotic prophase I. CHEK1 promotes the phosphorylation of CDC25A protein phosphatase to delay cell cycle progression in response to double-stranded DNA breaks. High expression of CHEK1 was associated with poor clinical characteristics of multiple myeloma patients (31). CHEK1 inhibitors have been shown to potentiate in combination with chemotherapy and their single-agent antitumor, in particular gemcitabine (32-35). Studies indicated the therapeutic effects of CHEK1 inhibition are related to p53-deficiency (36).
Thymidylate synthase (TYMS) catalyzes the methylation of deoxyuridylate to deoxythymidylate. The function of TYMS is to maintain the dTMP (thymidine-5-prime monophosphate) pool critical for DNA replication and repair. TYMS has been a target for cancer chemotherapeutic agents, like 5-fluoro-2-prime-deoxyuridine, 5-fluorouracil, and some folate analogs. Some studies indicated that TYMS variants were associated with high-dose methotrexate in childhood acute lymphoblastic leukemia, severe hand-foot-syndrome, the risk of persistence of pre-neoplastic cervical lesions and the risk of head and neck cancer (37-40). TYMS levels associated with prognosis and chemotherapy response drives the phenotypes of epithelial-tomesenchymal transition in NSCLC. The results established TYMS as a theranostic NSCLC marker related with survival, chemo-resistance and epithelial-to-mesenchymal transition (41,42).
The serpin family B member 5 (SERPINB5), located on chromosome 18q21.33 as a tumor suppressor gene, plays a critical in cancer cell invasion and metastasis. SERPINB5 variants are significantly associated with gallbladder cancer risk (43). Upregulated Maspin inhibited the expression of IKKα to promote cell apoptosis and delayed the development of the precancerous lesions in precancerous rats (44). The high expression of SERPINB5 significantly increased the recurrence rate and shortened disease-free survival in patients with oral squamous cell carcinoma (45). Cytoplasmic immunoreactive scores results showed that the expression of SERPINB5 was significantly higher in cervical cancer patients compared to cervical intraepithelial neoplasia. These studies indicated SERPINB5 was related to the survival in cervical cancer, pancreatic ductal adenocarcinoma and oral squamous cell carcinoma (45-47). Wang et al. reported that SERPINB5 had a statistically negative correlation with NSCLC prognosis and might be a promising prognostic signature in NSCLC (48).
The above studies have reported that the 4 hub DEGs (RRM2, CHEK1, TYMS and SERPINB5) were closely related to progression and prognosis of different cancers. A small number of studies have demonstrated that above 4 hub DEGs enriched in the p53 signaling or pyrimidine metabolism pathways play a vital role in lung cancer and overall survival of NSCLC patients. The current study also has several limitations. Firstly, there is not survival information for SCLC patients currently available in the online Kaplan-Meier plotter and GEPIA database. So, survival analysis has been not carried out for patients with SCLC. Secondly, we only assess the prognostic value and may miss some valuable information, which lacked of more clinical characteristics information from public databases, such as age, sex and treatment. Thirdly, our findings lacked molecular biological experimental validation of hub DEGs in NSCLC or SCLC.
Conclusions
Our study identified 4 hub DEGs (RRM2, CHEK1, TYMS and SERPINB5) in SCLC&NSCLC tissues compared with normal tissues. Functional analysis results indicated that these DEGs had different biological functions and were significantly enriched in different pathways. RRM2, CHEK1, TYMS and SERPINB5, which are mainly enriched in the p53 signaling and pyrimidine metabolism pathway, were significantly associated with the overall survival of NSCLC patients. These genes and pathways could serve as potential prognostic markers for personalized oncology in NSCLC or SCLC. However, more basic experiments and molecular mechanisms are needed to be confirmed for clinical applications.
Acknowledgments
Funding: This research was supported by startup funding for scientific research from Fujian Medical University (No. 2017XQ1216) and Research Project of Science and Technology Innovation Think Tank from Fujian Association for Science and Technology (No. FJKX-B2007).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-245/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-245/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-245/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Howlader N, Forjaz G, Mooradian MJ, et al. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N Engl J Med 2020;383:640-9. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Li D, Yin Y, He M, et al. Identification of Potential Biomarkers Associated with Prognosis in Gastric Cancer via Bioinformatics Analysis. Med Sci Monit 2021;27:e929104. [Crossref] [PubMed]
- Giannos P, Kechagias KS, Gal A. Identification of Prognostic Gene Biomarkers in Non-Small Cell Lung Cancer Progression by Integrated Bioinformatics Analysis. Biology (Basel) 2021;10:1200. [Crossref] [PubMed]
- Taniguchi H, Sen T, Rudin CM. Targeted Therapies and Biomarkers in Small Cell Lung Cancer. Front Oncol 2020;10:741. [Crossref] [PubMed]
- Zengin T, Önal-Süzek T. Comprehensive Profiling of Genomic and Transcriptomic Differences between Risk Groups of Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. J Pers Med 2021;11:154. [Crossref] [PubMed]
- Wu J, Hao Z, Ma C, et al. Comparative proteogenomics profiling of non-small and small lung carcinoma cell lines using mass spectrometry. PeerJ 2020;8:e8779. [Crossref] [PubMed]
- Yue C, Ma H, Zhou Y. Identification of prognostic gene signature associated with microenvironment of lung adenocarcinoma. PeerJ 2019;7:e8128. [Crossref] [PubMed]
- Šutić M, Vukić A, Baranašić J, et al. Diagnostic, Predictive, and Prognostic Biomarkers in Non-Small Cell Lung Cancer (NSCLC) Management. J Pers Med 2021;11:1102. [Crossref] [PubMed]
- Tantai JC, Pan XF, Zhao H. Network analysis of differentially expressed genes reveals key genes in small cell lung cancer. Eur Rev Med Pharmacol Sci 2015;19:1364-72. [PubMed]
- Liao Y, Yin G, Wang X, et al. Identification of candidate genes associated with the pathogenesis of small cell lung cancer via integrated bioinformatics analysis. Oncol Lett 2019;18:3723-33. [Crossref] [PubMed]
- Li X, Ma C, Luo H, et al. Identification of the differential expression of genes and upstream microRNAs in small cell lung cancer compared with normal lung based on bioinformatics analysis. Medicine (Baltimore) 2020;99:e19086. [Crossref] [PubMed]
- Ni M, Liu X, Wu J, et al. Identification of Candidate Biomarkers Correlated With the Pathogenesis and Prognosis of Non-small Cell Lung Cancer via Integrated Bioinformatics Analysis. Front Genet 2018;9:469. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J 2021;19:4101-9. [Crossref] [PubMed]
- Kretschmer C, Sterner-Kock A, Siedentopf F, et al. Identification of early molecular markers for breast cancer. Mol Cancer 2011;10:15. [Crossref] [PubMed]
- Grade M, Hummon AB, Camps J, et al. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int J Cancer 2011;128:1069-79. [Crossref] [PubMed]
- Wang J, Yi Y, Chen Y, et al. Potential mechanism of RRM2 for promoting Cervical Cancer based on weighted gene co-expression network analysis. Int J Med Sci 2020;17:2362-72. [Crossref] [PubMed]
- Wang S, Wang XL, Wu ZZ, et al. Overexpression of RRM2 is related to poor prognosis in oral squamous cell carcinoma. Oral Dis 2021;27:204-14. [Crossref] [PubMed]
- Mazzu YZ, Armenia J, Nandakumar S, et al. Ribonucleotide reductase small subunit M2 is a master driver of aggressive prostate cancer. Mol Oncol 2020;14:1881-97. [Crossref] [PubMed]
- Han J, Hu J, Sun F, et al. MicroRNA-20a-5p suppresses tumor angiogenesis of non-small cell lung cancer through RRM2-mediated PI3K/Akt signaling pathway. Mol Cell Biochem 2021;476:689-98. [Crossref] [PubMed]
- Jin CY, Du L, Nuerlan AH, et al. High expression of RRM2 as an independent predictive factor of poor prognosis in patients with lung adenocarcinoma. Aging (Albany NY) 2020;13:3518-35. [Crossref] [PubMed]
- Liu XP, Huang Q, Yin XH, et al. Strong Correlation between the Expression of CHEK1 and Clinicopathological Features of Patients with Multiple Myeloma. Crit Rev Eukaryot Gene Expr 2020;30:349-57. [Crossref] [PubMed]
- Karp JE, Thomas BM, Greer JM, et al. Phase I and pharmacologic trial of cytosine arabinoside with the selective checkpoint 1 inhibitor Sch 900776 in refractory acute leukemias. Clin Cancer Res 2012;18:6723-31. [Crossref] [PubMed]
- Daud AI, Ashworth MT, Strosberg J, et al. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as monotherapy and in combination with gemcitabine in patients with advanced solid tumors. J Clin Oncol 2015;33:1060-6. [Crossref] [PubMed]
- Walton MI, Eve PD, Hayes A, et al. CCT244747 is a novel potent and selective CHK1 inhibitor with oral efficacy alone and in combination with genotoxic anticancer drugs. Clin Cancer Res 2012;18:5650-61. [Crossref] [PubMed]
- Xiao Y, Ramiscal J, Kowanetz K, et al. Identification of preferred chemotherapeutics for combining with a CHK1 inhibitor. Mol Cancer Ther 2013;12:2285-95. [Crossref] [PubMed]
- Ma CX, Cai S, Li S, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest 2012;122:1541-52. [Crossref] [PubMed]
- Al-Sheikh A, Yousef AM, Alshamaseen D, et al. Effects of thymidylate synthase polymorphisms on toxicities associated with high-dose methotrexate in childhood acute lymphoblastic leukemia. Cancer Chemother Pharmacol 2021;87:379-85. [Crossref] [PubMed]
- Silva NNT, Santos ACS, Nogueira VM, et al. 3'UTR polymorphism of Thymidylate Synthase gene increased the risk of persistence of pre-neoplastic cervical lesions. BMC Cancer 2020;20:323. [Crossref] [PubMed]
- Hamzic S, Kummer D, Froehlich TK, et al. Evaluating the role of ENOSF1 and TYMS variants as predictors in fluoropyrimidine-related toxicities: An IPD meta-analysis. Pharmacol Res 2020;152:104594. [Crossref] [PubMed]
- De Castro TB, Rodrigues-Fleming GH, Oliveira-Cucolo JG, et al. Gene Polymorphisms Involved in Folate Metabolism and DNA Methylation with the Risk of Head and Neck Cancer. Asian Pac J Cancer Prev 2020;21:3751-9. [Crossref] [PubMed]
- Siddiqui MA, Gollavilli PN, Ramesh V, et al. Thymidylate synthase drives the phenotypes of epithelial-to-mesenchymal transition in non-small cell lung cancer. Br J Cancer 2021;124:281-9. [Crossref] [PubMed]
- Agulló-Ortuño MT, García-Ruiz I, Díaz-García CV, et al. Blood mRNA expression of REV3L and TYMS as potential predictive biomarkers from platinum-based chemotherapy plus pemetrexed in non-small cell lung cancer patients. Cancer Chemother Pharmacol 2020;85:525-35. [Crossref] [PubMed]
- Mahananda B, Vinay J, Palo A, et al. SERPINB5 Genetic Variants rs2289519 and rs2289521 are Significantly Associated with Gallbladder Cancer Risk. DNA Cell Biol 2021;40:706-12. [Crossref] [PubMed]
- Wang N, Chang LL. The potential function of IKKα in gastric precancerous lesion via mediating Maspin. Tissue Cell 2020;65:101349. [Crossref] [PubMed]
- Kawasaki M, Sakabe T, Kodani I, et al. Cytoplasmic-only Expression of Maspin Predicts Poor Prognosis in Patients With Oral Squamous Cell Carcinoma. Anticancer Res 2021;41:4563-70. [Crossref] [PubMed]
- Isci Bostanci E, Guler I, Dikmen AU, et al. Prognostic role of maspin expression in patients with cervical dysplasia and cervical cancer. J Obstet Gynaecol Res 2020;46:759-64. [Crossref] [PubMed]
- Uchinaka EI, Sakabe T, Hanaki T, et al. Cytoplasmic-only Expression of Maspin Predicts Unfavorable Prognosis in Patients With Pancreatic Ductal Adenocarcinoma. Anticancer Res 2021;41:2543-52. [Crossref] [PubMed]
- Wang XF, Liang B, Zeng DX, et al. The roles of MASPIN expression and subcellular localization in non-small cell lung cancer. Biosci Rep 2020;40:BSR20200743. [Crossref] [PubMed]


